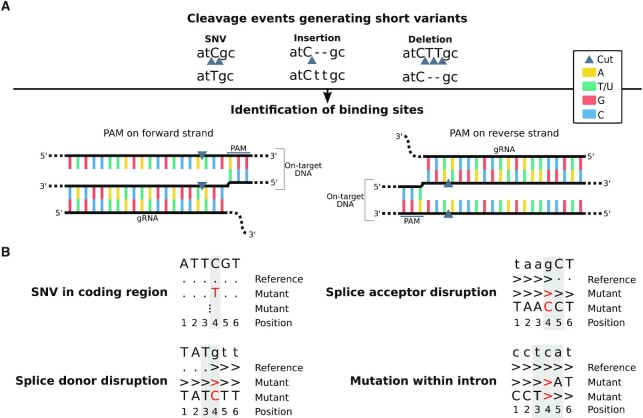
Figure 2.
Analysis of sequence variations at possible on-/off-targets. (A) Strategy for variant-based off-target screening. Short genomic variants discovered from RNA-seq are screened to find Cas9 binding sites proximal to the possible ‘cut’ positions associated to the variants. All gRNA–DNA interactions ending at one of the identified binding sites are evaluated, and the energetically most favourable one is retained as most likely off-target for each variant. (B) Patterns of on-target single nucleotide variations. Four different types of on-target editing events are shown. For each of them, the reference pileup and examples of other possible mutant pileups (in red) are given. The positions analyzed to evaluate on-target edits are highlighted with grey boxes.