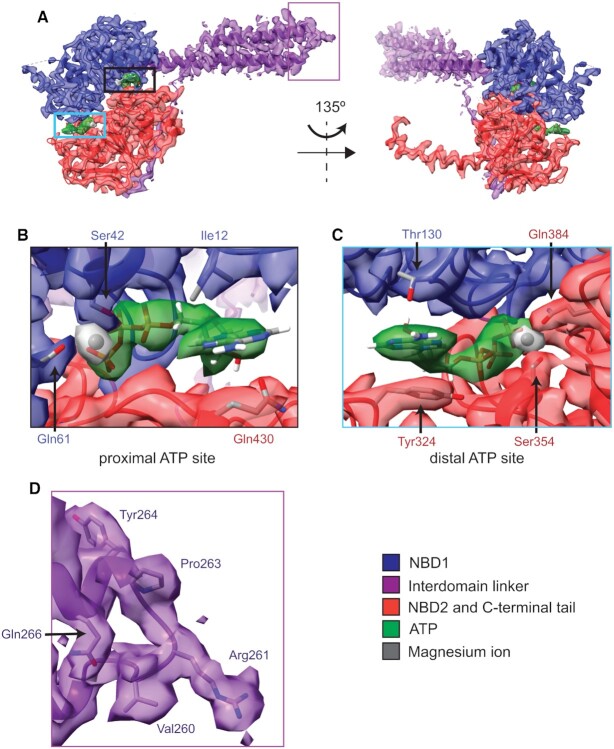
Figure 5.
Structure of Sal(B), focussing on ATP-binding sites and the interdomain linker loop. (A) Atomic model and cryo-EM density of Sal(B), showing NBD1 (blue), the interdomain linker (purple), NBD2 and the C-terminal tail (red), two sandwiched ATP molecules (green) and magnesium ions that coordinate the β- and γ-phosphates of ATP (grey). Left, front view; right, back view. (B) Proximal ATP binding site showing atomic models of the ATP molecule, magnesium ion and selected Sal(B) protein side chains involved in ATP binding. (C) Distal ATP binding site showing atomic models of the ATP molecule, magnesium ion and selected Sal(B) protein side chains involved in ATP binding. (D) The loop between the two helices of the interdomain linker. The density of this region is well resolved, allowing for unambiguous visualisation of side chains.