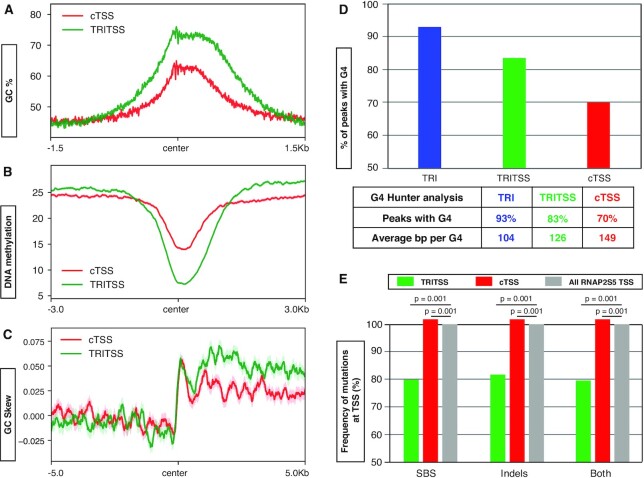
Figure 5.
Sequences and mutations at TRIs. (A) Median signal profile of percent GC of the mm10 mouse genome centered near the TSSs of TRITSSs (green) and cTSSs (red). (B) Median DNA methylation signal profile of mouse B cells centered near the TSSs of TRITSSs (green) and cTSSs (red). For TRIs versus cTSS, P < 1.0 × 10–250 and TRITSS versus cTSS, P = 8.3 × 10–238; Wilcoxon rank sum test. (C) Median signal profile of GC skew of the mm10 mouse genome centered near the TSSs of TRITSSs (green) and cTSSs (red). GC skew of TRIs and TRITSS are significantly higher than cTSS (P = 1.17 × 10–66 and P = 1.48 × 10–67 respectively, Wilcoxon rank sum test). (D) Table and bar graph of the predicted frequency of G-quadruplex formation at TRIs, TRITSSs and cTSSs. (E) Bar graph of single basepair substitutions (SBS) and short insertions/deletions (indels) at TRIs, TRITSSs and cTSSs normalized to cTSSs showing empirical P-value. *Shaded areas on all unfilled line plots are the standard error.