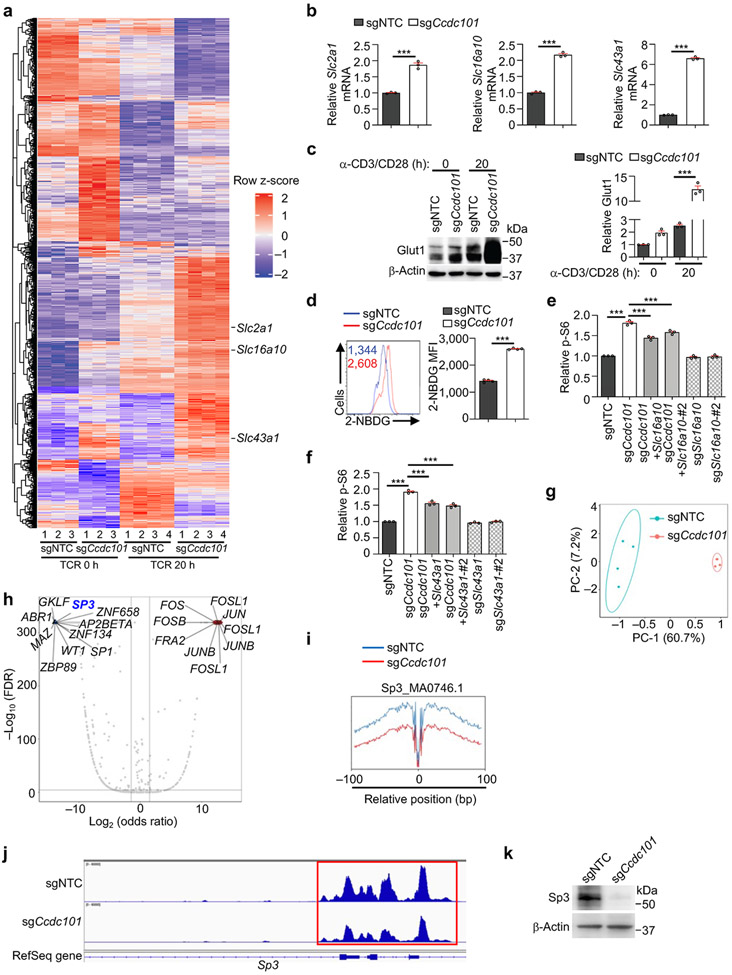
Extended Data Figure 9 (to Figure 4). SAGA complex represses the expression of nutrient transporters and mTORC1 activation.
(a) Heatmap of differentially expressed genes in Ccdc101-null Treg cells stimulated with α-CD3/CD28 antibodies for 0 (n = 3 samples each group) or 20 h (n = 4 samples each group). (b) Slc2a1, Slc16a10 or Slc43a1 mRNA expression in sgNTC- or sgCcdc101-transduced cells at steady state (n = 3 samples each group). (c) Immunoblot analysis and quantification of relative Glut1 expression in sgNTC- or sgCcdc101-transduced cells that were stimulated with α-CD3/CD28 antibodies for 0 or 20 h (n = 3 samples each group). (d) Flow cytometry analysis and quantification of 2-NBDG uptake in sgNTC or sgCcdc101 (both Ametrine+)-transduced cells were stimulated with α-CD3/CD28 antibodies for 20 h (n = 4 samples each group). (e, f) Quantification of relative p-S6 level in cells transduced with the indicated sgRNAs that were stimulated with TCR for 3 h (n = 3 samples each group). (g) Principal component analysis (PCA) of ATAC-seq for cells transduced with sgNTC (n = 4 samples) or sgCcdc101 (both Ametrine+) (n = 3 samples) and stimulated with α-CD3/CD28 antibodies for 20 h. (h) Motif enrichment analysis of ATAC-seq of sgNTC- and sgCcdc101-transduced cells (n = 4 samples each group). (i) Footprinting analysis of Sp3 binding in ATAC-seq. (j) Accessibility of the Sp3 locus in sgNTC- and sgCcdc101-transduced cells as identified by ATAC-seq. Highlighted peaks in the red box indicate differential accessible regions. (k) Immunoblot analysis of Sp3 expression in sgNTC- or sgCcdc101 (both Ametrine+)-transduced cells. Mean ± s.e.m. (b–f). ***P < 0.001; two-tailed unpaired Student’s t-test (b, d); one-way ANOVA (c, e, f). Data are representative of one (a, g–j), two (b, e, f, k), or pooled from two (c, d) experiments.