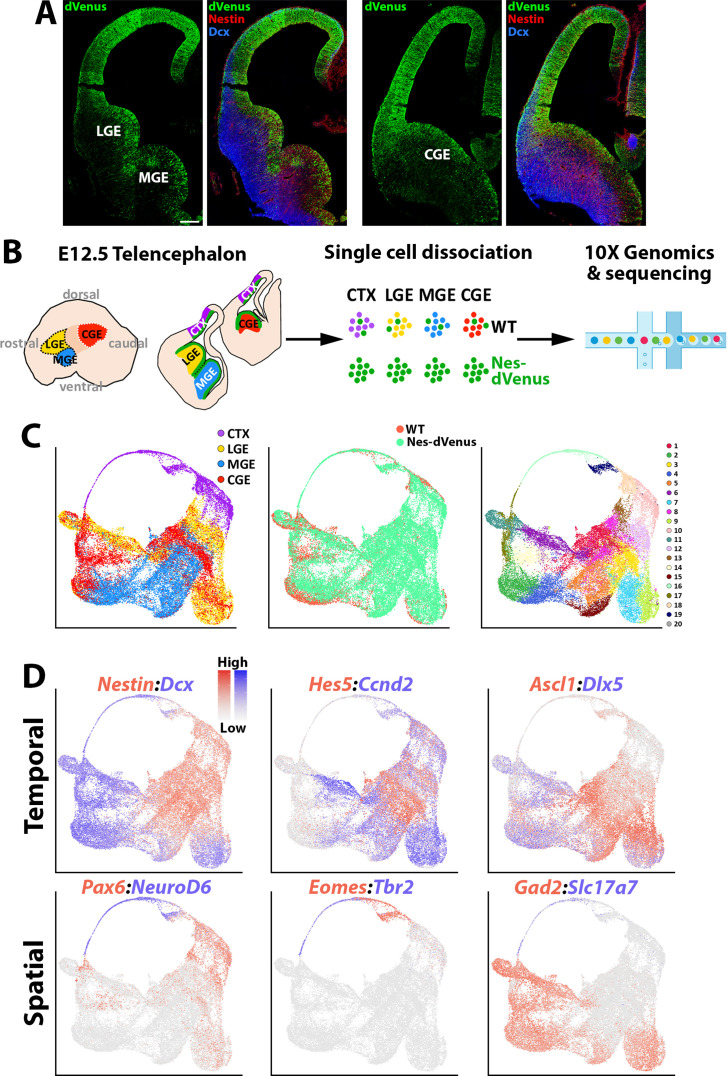
Figure 1. scRNAseq of distinct E12.5 forebrain regions from WT and Nes-dVenus mice.
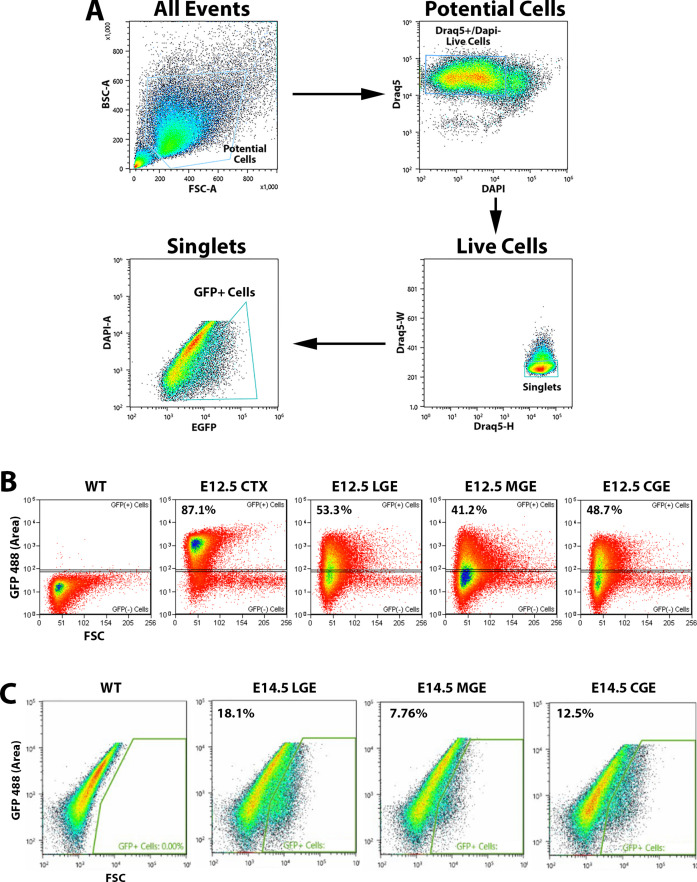
(A) Coronal section through the LGE/MGE (left) and CGE (right) of a E12.5 Nes-dVenus telencephalon immunostained for GFP (green), Nestin (Red) and Dcx (Blue). Scale bar = 200 μm. (B) Schematic of telencephalic cell dissection and single-cell dissociation. (C) Uniform manifold approximation and projection (UMAP) plots of all cells labeled by brain region (left), mouse line (middle) or putative cell clusters (right). (D) Representative genes displaying enriched temporal and/or spatial expression patterns. CTX, cortex; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; WT, wild-type.