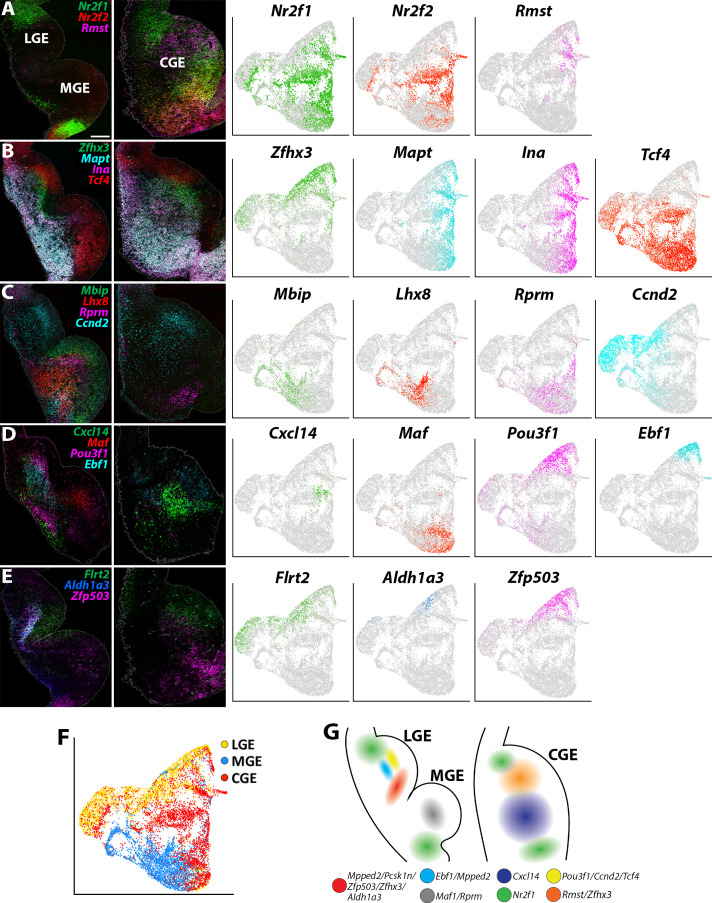
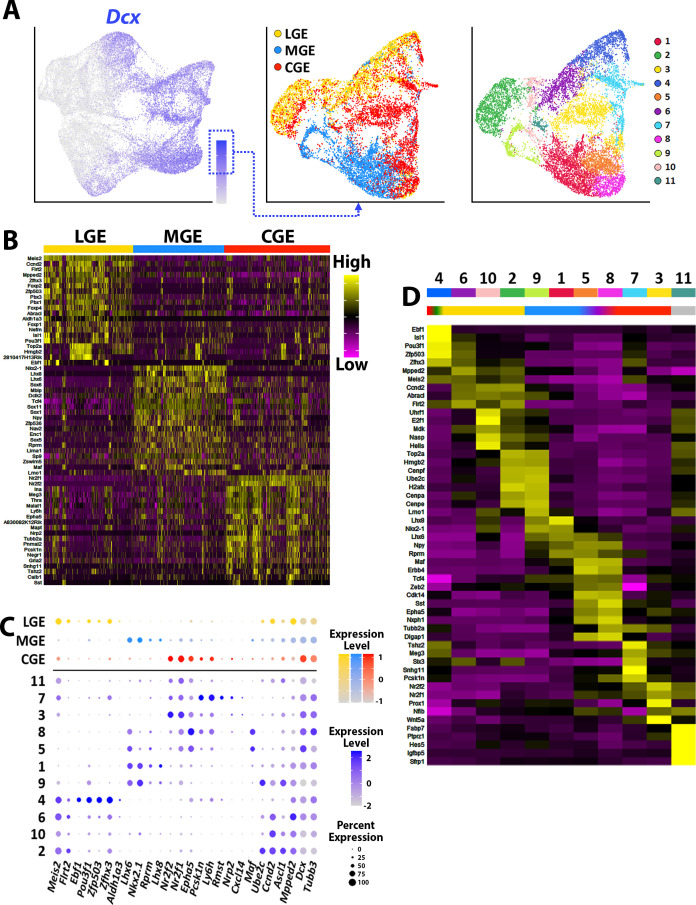
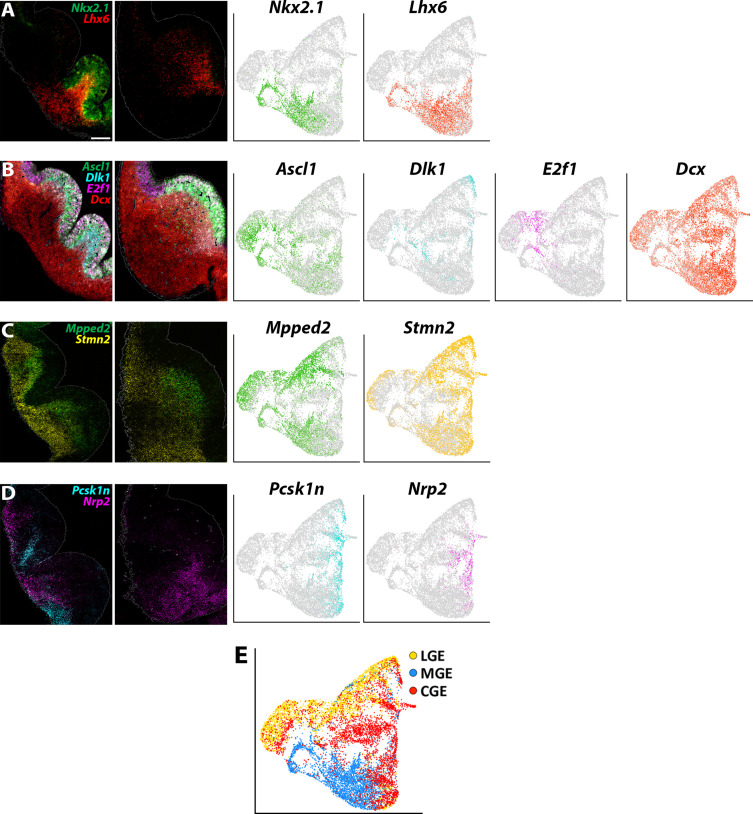
Figure 6. Expression profiles of spatially-enriched genes in SVZ/MZ cells in the ganglionic eminences.
(A–E) Left, RNAscope HiPlex and HiPlexUp in situ hybridization assays of differentially expressed SVZ/MZ genes on E12.5 GE coronal sections. Right, the corresponding UMAP plots of SVZ/MZ-enriched genes depicted in the in situs. Scale bar in A = 200 μm. (F) UMAP plot of Dcx-enriched cells annotated by brain region. (G) Schematic of a coronal section through the E12.5 ventral telencephalon depicting the approximate spatial location of SVZ/MZ-enriched genes within the GEs.