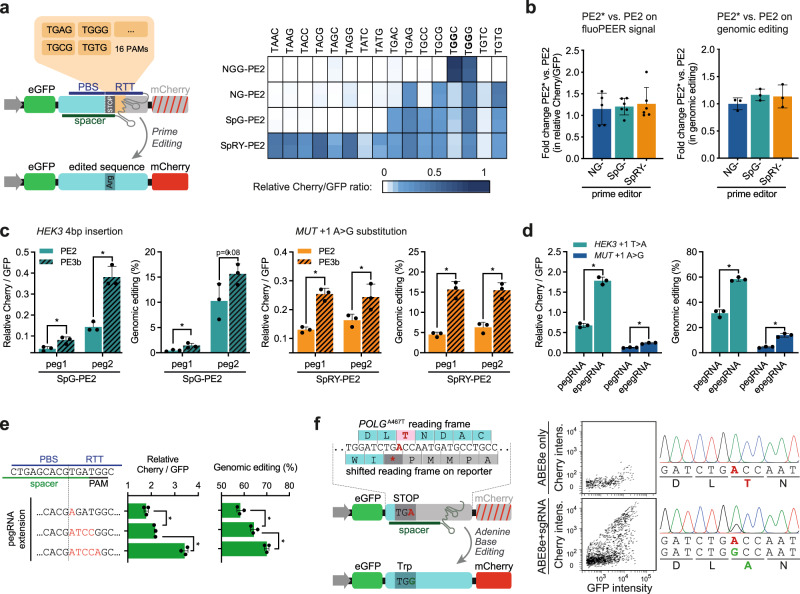
Fig. 2. FluoPEER enables characterization of various genome editing techniques.
a 16 fluoPEER plasmids were constructed to contain the same pegRNA spacer binding site (HEK3) including a stop codon, followed by 16 different 4-nucleotide PAM sequences. Prime editing using a pegRNA that converts this stop codon to an arginine-encoding codon results in Cherry signal. The heatmap shows the fluoPEER signal for these 16 plasmids, four prime editor (PE2) variants, and replicates (n = 2) per condition in HEK293T cells. b Prime editor variants with adapted nuclear localization sequences (PE2*) were tested in HEK293T cells. Graphs show a summary of all fluoPEER scores (left panel) and of the conversion efficiency of the genomic mutation (right panel), expressed as the fold change (FC) of the PE2* variants relative to the corresponding PE2 variants. See also Supplementary Fig. 8. n = 2–3 biologically independent replicates. c FluoPEER was used to evaluate nicking sgRNAs for the PE3b technique in HEK293T cells. PE3b designs increased fluoPEER scores (Cherry/GFP ratios) as well as genomic editing as measured by NGS. d Improved prime editing using epegRNAs was measured on fluoPEER. Cherry over GFP was measured on fluoPEER; genomic editing was measured using NGS. e Increased mismatches between the pegRNA RTT and the target sequence resulted in higher prime editing efficiency on fluoPEER and the genome. Three pegRNAs with varying mismatches with the genome were adopted from Chen et al.5. More mismatches resulted in higher editing efficiency on the genome and fluoPEER. Significance was analyzed using a two-tailed unpaired Student’s t test (*P < 0.05) for n = 3 biologically independent replicates for c–e. f FluoPEER can report base editing when editing of the target nucleotide resolves a stop codon in any possible reading frame. The genomic region of the POLGA467T mutation was inserted into fluoPEER with a shifted reading frame to create a stop codon (left). This fluoPEER was transfected into fibroblasts with biallelic POLGA467T mutations to show base editing on the reporter (middle) and the genome (right). Error bars represent standard deviations from the mean. Source data are provided as a Source Data file.