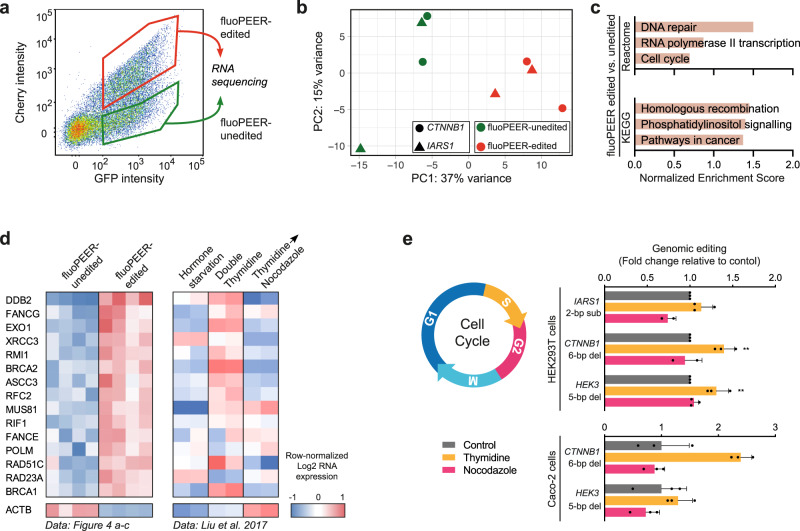
Fig. 4. Endogenous DNA repair proteins and the cell cycle affect prime editing outcomes.
a Schematic overview of RNA-sequencing set-up. FluoPEER-edited and -unedited HEK293T cells were sorted separately and RNA was sequenced. b PCA plot and c gene set enrichment analysis of RNA sequencing of the fluoPEER-edited and -unedited cell populations show expression-based differences in DNA repair- and cell cycle-associated genes. d Left panel shows a heatmap of the expression of DNA-repair-associated genes which were enriched in the transcriptome of fluoPEER-edited vs. fluoPEER-unedited HEK293T cells from a–c. Right panel shows expression of the same genes in publicly available transcriptome profiles (GSE94479) of MCF-7 cells stalled in different cell cycle phases49. For both transcriptomic datasets, log2-transformed expression values were mean centered per gene for visualization. e Cell cycle synchronization at the G1/S boundary (double thymidine block) or G2/M phase (nocodazole block) affects genomic prime editing efficiency in HEK293T and Caco-2 cells. HEK293T replicates were normalized to the average editing of the control condition for each of three repeated, independent experiments; Caco-2 replicates (n = 3) were normalized to the average editing of the control condition for a single representative experiment with three biological replicates. Error bars represent standard deviations from the mean. Source data are provided as a Source Data file.