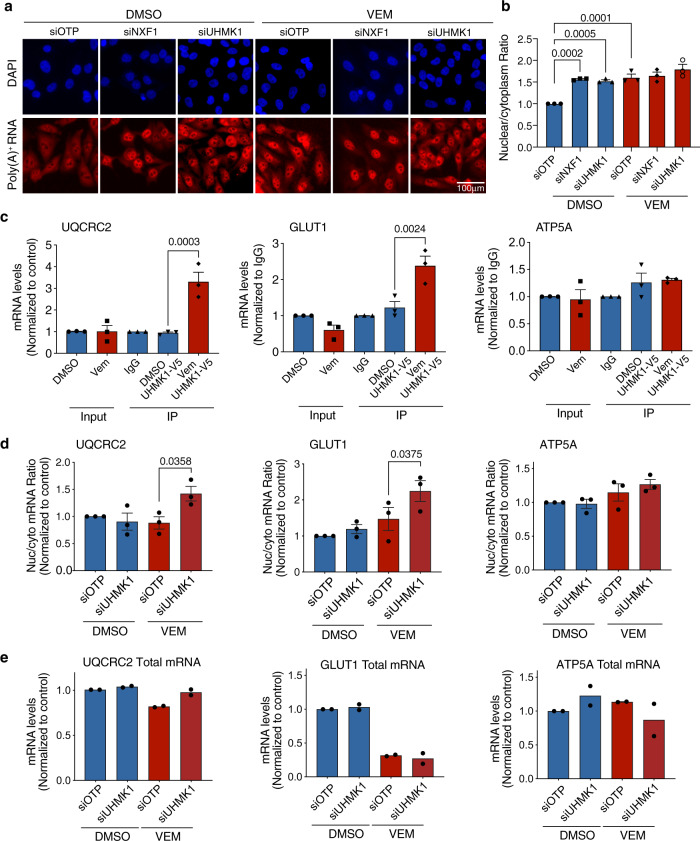
Fig. 5. UHMK1 associates with mRNA encoding metabolic proteins and promotes selective mRNA transport in BRAFV600 melanoma cells adapting to BRAFi.
A375 cells were transfected with the indicated siRNA and treated with DMSO or 1 μM Vem for 48 h. a RNA fluorescence in situ hybridization (FISH) using a poly(A)+ RNA specific probe in A375 cells treated as indicated (representative of n = 3 biologically independent experiments). b The nuclear to cytoplasm ratio of poly(A)+ RNA was calculated using high content image analysis. Data are expressed as mean fold change ± SEM (n = 3 biologically independent experiments). Statistical significance was determined using a one-way ANOVA adjusted for multiple comparisons. c RNA immunoprecipitation (RNA-IP) assays were performed in UHMK1-V5 expressing A375 cells following treatment with DMSO or 1 μM Vem for 48 h. The indicated mRNA transcripts were then analysed using RT-qPCR. Data are expressed as mean ± SEM (n = 3 biologically independent experiments). Statistical significance was determined using a one-way ANOVA adjusted for multiple comparisons. d Cell lysates were fractionated into nuclear and cytoplasmic pools of RNA and analysed for the indicated genes using RT-qPCR. The nuclear/cytoplasm (Nuc/Cyto) ratio was calculated from analysis of individual nuclear and cytoplasmic compartments. Data are expressed as mean ± SEM (n = 3 biologically independent experiments). See also Supplementary Fig. 5C. Statistical significance was determined using a one-way ANOVA adjusted for multiple comparisons. e Whole-cell lysates were used to assess total mRNA levels. Data are expressed as mean ± SEM (n = 2 biologically independent experiments). See also Supplementary Fig. 5. Source data are provided as a Source Data file.