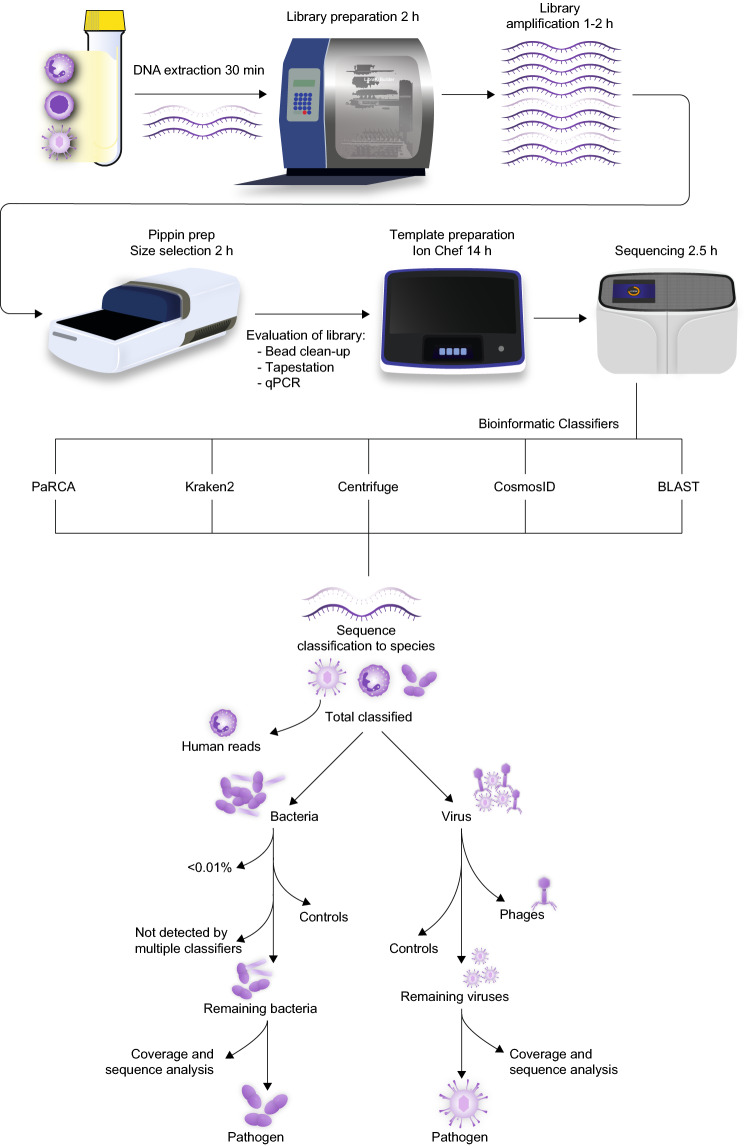
Figure 1.
DNA metagenomic sequencing workflow. DNA from cerebrospinal fluid specimens, containing leukocytes and pathogens, was extracted and followed by library preparation and sequencing. Datasets generated by the Ion S5 were processed by four different bioinformatics classifiers to profile the microbiome. BLAST was used for verification. Flowchart for identification of pathogens by removing false positive species. Virus contaminants can be removed by comparison of sample datasets with controls by which environmental and bioinformatic misclassifications are identified. Phages can be disregarded as these viruses do not infect human cells. A final manual examination of remaining viral reads is required for coverage and sequence analysis. The bacterial contaminants were removed by applying a filter of cutoff value and comparison between classifiers and controls followed by a manual examination.