Figure 5.
Molecular determinants of MATR3 toxicity, aggregation, and localization
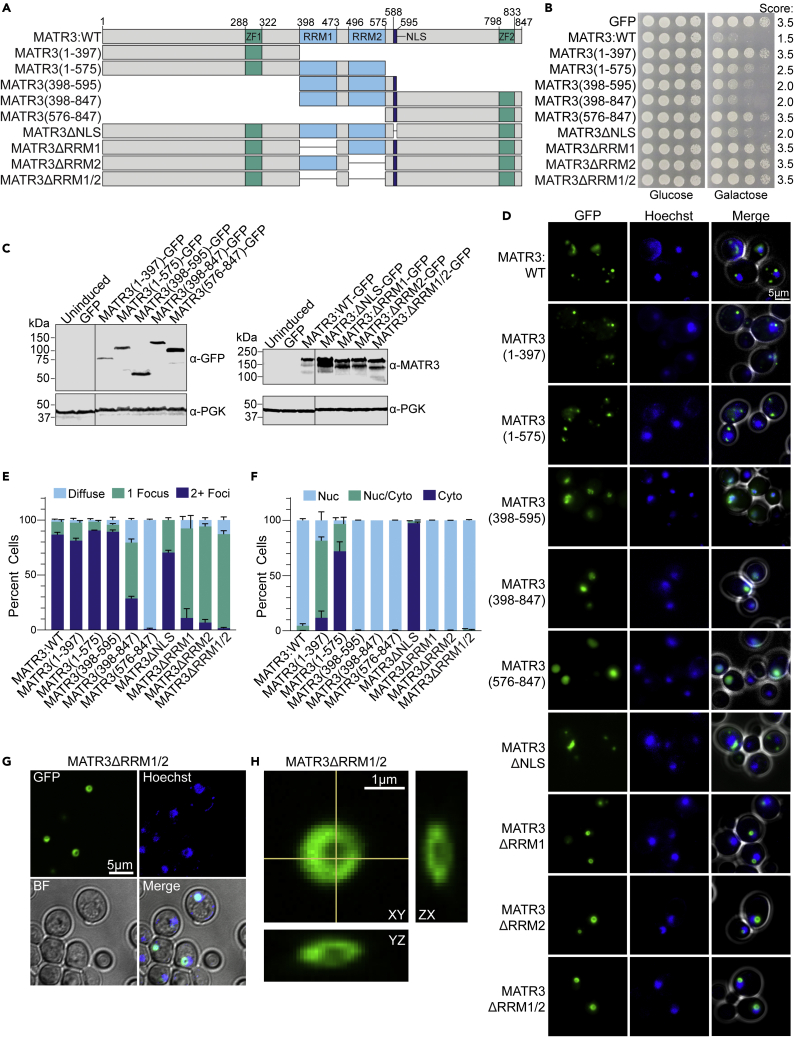
(A) MATR3 domain map showing the constructs used in this study.
(B) W303Δhsp104 yeast was transformed with the indicated pAG423GAL-MATR3-GFP constructs or vector control. Strains were serially diluted 5-fold and spotted in duplicate on glucose (non-inducing) and galactose (inducing) media. Scores are based on number of spots from an average of at least three replicates.
(C) Strains from B were induced for 5h, lysed, and immunoblotted for MATR3, GFP (mouse), and PGK (loading control). Immunoblots were cropped to remove additional lanes for presentation purposes, and crop lines are marked with a line.
(D) Strains from B were induced for 5 h, stained with Hoechst dye, and imaged. Representative images are shown. Merge channel includes GFP, Hoechst, and bright field images. Scale bar, 5μm.
(E and F) Quantification of microscopy experiments shown in D. The percentages of cells with diffuse (light blue), one focus (teal), or multiple foci (dark blue) are quantified (E). The percentages of cells with nuclear (Nuc, light blue), nuclear and cytoplasmic (Nuc/Cyto, teal), or cytoplasmic (Cyto, dark blue) localization of MATR3-GFP is quantified (F). N ≥ 3, with approximately 100 cells quantified per trial. Error bars show mean ± SEM.
(G) pAG423GAL-MATR3ΔRRM1/2-GFP yeast were induced for 5 h, stained with Hoechst dye, and imaged by confocal microscopy. A representative z slice is shown. Scale bar, 5μm.
(H) Expanded orthogonal view of a focus from the same strain depicted in (G) Scale bar, 1μm. See also Figure S2.