Figure 2.
Computationally identified mesenchymal clusters represent spatially distinct populations
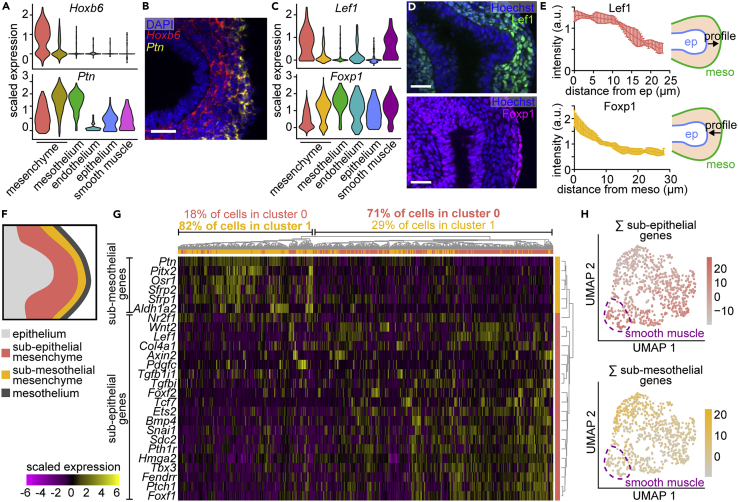
(A) Violin plots showing the expression of Hoxb6 or Ptn in each cluster.
(B) Fluorescence in situ hybridization for Hoxb6 and Ptn in E11.5 lungs. Scale bar shows 25 μm.
(C) Violin plots showing the expression of Lef1 or Foxp1 in each cluster.
(D) E11.5 lungs immunostained for cluster 0 marker Lef1 or for cluster 1 marker Foxp1 and counterstained with Hoechst. Scale bars show 25 μm.
(E) Quantifications of Lef1 and Foxp1 intensity profiles emanating from the epithelium (for Lef1) or from the mesothelium (for Foxp1). Schematics show lines and direction along which intensity profiles were measured. Mean and SD are plotted (n = 4).
(F) Schematic depicting the sub-epithelial and sub-mesothelial compartments of the mesenchyme.
(G) Heatmap showing the expression of genes specific to either mesenchymal compartment. Genes (rows) are clustered based on the dendrogram to the right. Cells (columns) are clustered based on the dendrogram above, and each column is color-coded according to the original cluster identity from Figure 1B.
(H) UMAP of mesenchymal and smooth muscle cells color-coded according to the sum of their expression of either sub-epithelial or sub-mesothelial mesenchyme marker genes. Dotted line indicates the location of smooth muscle cells