Figure 5.
Diffusion analysis reveals differentiation trajectory of mesenchymal cells into airway smooth muscle
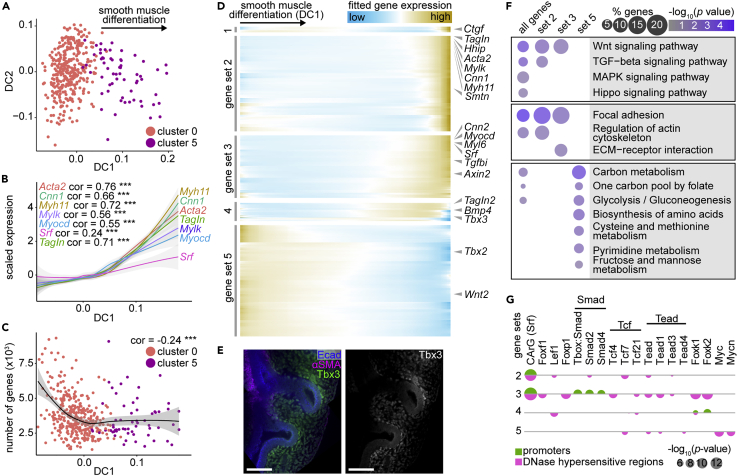
(A) Diffusion plots of sub-epithelial mesenchymal and smooth muscle cells.
(B) Scaled expression of known smooth muscle markers versus cell loadings along DC1. Pearson correlation coefficients and significance are indicated on plots and lines represent smoothed data with SE shaded in gray. ∗∗∗ indicates p< 0.0001.
(C) Number of genes detected per cell versus DC1. Pearson correlation coefficients and significance are indicated on plots and lines represent smoothed data with SE shaded in gray. ∗∗∗ indicates p< 0.0001.
(D) Heatmap showing five gene sets identified using hierarchical agglomerative clustering of spline-fitted gene-expression profiles along DC1. Genes of interest are indicated to the right.
(E) E11.5 lungs immunostained for Tbx3 (gene set 4), Ecad, and αSMA. Scale bars show 50 μm.
(F) Bubble plot showing the enrichment percentage and adjusted p value of relevant KEGG pathways for the gene sets defined in (E)
(G) Bubble plot showing the p values of motifs for transcription factor-binding sites in the promoters of and DNase-hypersensitive regions near genes of sets 2–5.