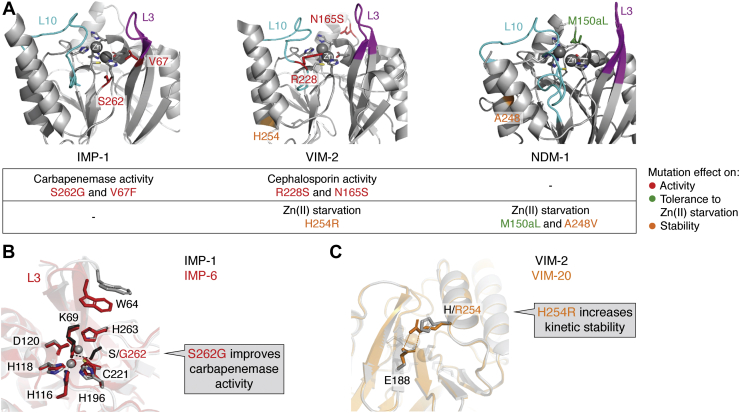
Figure 4.
A, key mutations and evolutionary traits in the main families of acquired MBLs: IMP-1, VIM-2, and NDM-1. Figures were rendered with PyMol based on the PDB IDs 4C1G, 4NQ2, and 3SPU, respectively. The active-site loops L10 and L3 are shown in cyan and magenta, respectively; the metal ligands as sticks, and the Zn(II) ions as spheres. The mutational “hot spots” are indicated in different colors according to their impact on catalytic activity (red) (79, 130, 139) or in the tolerance against Zn(II) starvation by improving metal binding (green) or the kinetic stability (orange) (73, 74, 79). B, a conformational change in loop L3 introduced by substitution S262G in IMP improves carbapenemase activity. The interaction between S262 and K69 marked with black dashes in IMP-1 (gray structure) is disrupted by the substitution in IMP-6 (red; PDB: 6LVJ). C, the substitution H254R increases the kinetic stability of VIM-20 (PDB: 6OP6) by the formation of a salt bridge between R254 and E188. IMP-1, imipenemase 1; MBL, metallo-β-lactamase; NDM-1, New Delhi metallo-β-lactamase 1; PDB, Protein Data Bank; VIM-2, Verona imipenemase 2.