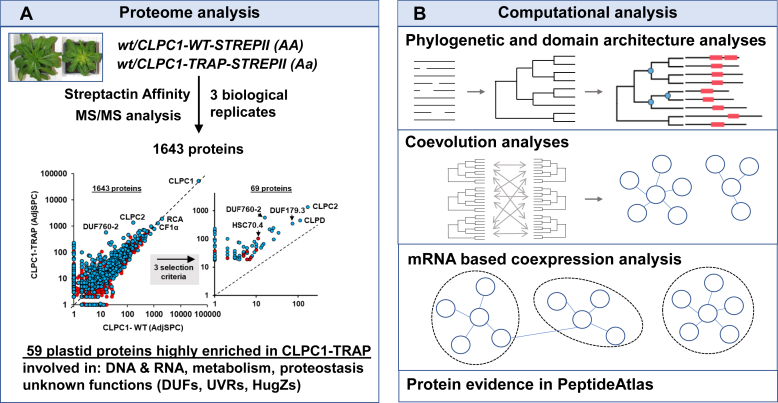
Figure 2.
Schematic overview of the proteome affinity analysis for CLPC1-WT and CLPC1-TRAP and four complementary bioinformatics analyses of selected proteins of interest.A, affinity protein enrichment using transgenic Arabidopsis plants expressing STREPII-tagged CLPC1-WT and CLPC1-TRAP with mutations in the Walker B motifs that block ATP hydrolysis. Proteins in the affinity eluates were identified and quantified by tandem mass spectrometry (MS/MS), and highly enriched proteins in the CLPC1-TRAP as compared with CLPC1-WT were selected for further analysis. The inset of the scatterplot was from Figure 1A. B, four complimentary bioinformatics analyses were carried out for a selected set of proteins of interest with unknown functions. These selected proteins are candidate adaptors for the CLP system. The phylogeny and domain analysis provide new evolutionary clues and information about possible redundancies. The ERC analysis aims to determine sign of coevolution of the proteins that make up the Clp system in chloroplasts and find coevolutionary signals between subunits of the Clp system and several of the enriched proteins. The mRNA-based coexpression analysis provides support for functional interactions between the Clp system and the enriched proteins. Mass spectrometry–based protein evidence across many public proteome datasets using the Arabidopsis PeptideAtlas database provided insight in the relative abundance and puts the enrichment in the CLPC1-TRAP in perspective. Because elucidation of Clp adaptor functions and even substrates can be so daunting, this comprehensive analysis of candidate adaptors and substrates allows for more rational choices of which proteins to select for functional studies and also help design the most promising experiments.