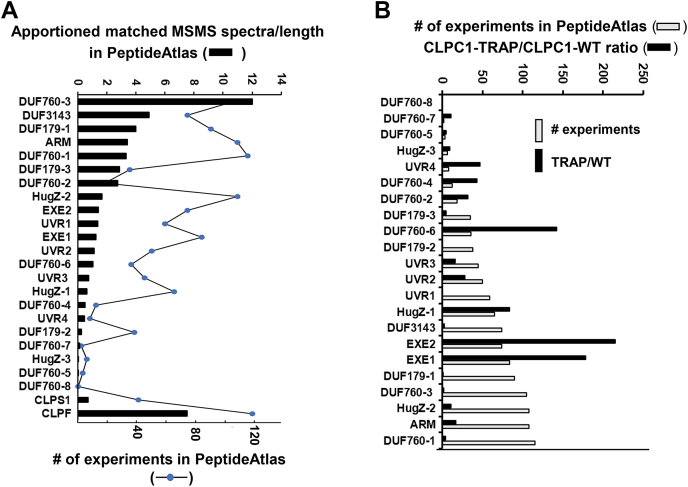
Figure 9.
Relative abundance and observations for proteins based on millions of MS/MS spectra in the Arabidopsis PeptideAtlas. Arabidopsis PeptideAtlas is based on publicly available MS data for a wide range of highly diverse samples from Arabidopsis (including leaves, flowers, roots, cell cultures, and subcellular fractions) and reanalyzed through a uniform processing and metadata annotation pipeline. The MS data of the CLPC1 trapping experiments, as described above, as well as our previous CLPC1 trapping study (11) are part of this atlas. A, relative protein abundance in the current Arabidopsis PeptideAtlas based on apportioned matched spectra (PSMS) per length in number of amino acids for the 22 proteins in Table 2, and for CLPS1 and CLPF. B, number of experiments in the current Arabidopsis PeptideAtlas for which each of the 22 proteins in Table 2 and CLPS1 and CLPF were observed to provide a rough measure of abundance across the many sample types. Also shown is the relative enrichment in the CLPC1-TRAP compared with CLPC1-WT affinity experiments. This illustrates that the enrichment is independent of the general cellular protein abundance and underscores that the ClpC1-TRAP is highly selective.