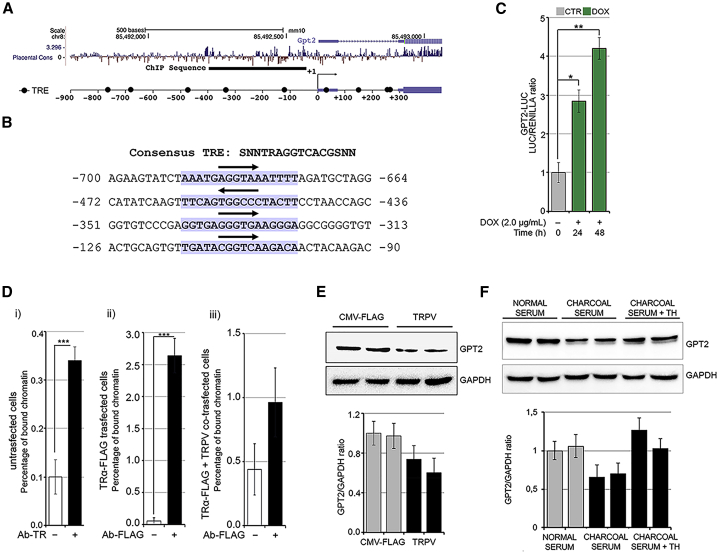
Figure 2.
Thyroid hormone positively regulates GPT2 transcription in muscle cells
(A) The mammalian conservation, the gene structure, and the position from Genome Browser are indicated for the GPT2 gene. The black bar represents the sequence analyzed by chromatin immunoprecipitation (ChIP). Black circles represent potential TH responsive elements (TREs) identified by in silico analysis. The transcription start site (TSS) is set at +1.
(B) Sequence of first four consensus TREs identified upstream of the TSS. Sequence matching the consensus is highlighted in purple, and the region corresponding to the conserved AGGTCA is indicated by an arrow.
(C) T3 transactivation activity was measured using the reporter plasmid GPT2-luciferase (LUC). C2C12 pTRE-D2 cultured with 2 μg/mL DOX for 24 and 48 h were transfected with the GPT2-LUC and CMV-Renilla as an internal CTR. The results are shown as means ± standard deviation (SD) of the LUC/Renilla ratios from at least 3 separate experiments, performed in duplicate.
(D) ChIP assay was performed in (1) C2C12 cells using the anti-TH receptor antibodies, (2) C2C12 cells transfected with TRα-FLAG using the anti-FLAG antibody, and (3) C2C12 cells co-transfected with TRα-FLAG + the dominant-negative of TH receptor (TRPV) using the anti-FLAG antibody.
(E) Protein levels of GPT2 were measured by western blot analysis in C2C12 cells transfected with TRPV or CMV-FLAG for 48 h.
(F) Protein levels of GPT2 were measured by western blot analysis in C2C12 cells cultured in normal serum, charcoal serum, and charcoal serum + TH for 48 h. The histograms below show the quantification of the GPT2 protein levels versus GAPDH levels. Data represent the mean ± SD of the mean of fifteen replicates. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.