Abstract
Giant unilamellar vesicles (GUVs) are frequently used as models of biological membranes and thus are a great tool to study membrane-related cellular processes in vitro. In recent years, encapsulation within GUVs has proven to be a helpful approach for reconstitution experiments in cell biology and related fields. It better mimics confinement conditions inside living cells, as opposed to conventional biochemical reconstitution. Methods for encapsulation inside GUVs are often not easy to implement, and success rates can differ significantly from lab to lab. One technique that has proven successful for encapsulating more complex protein systems is called continuous droplet interface crossing encapsulation (cDICE). Here, a cDICE-based method is presented for rapidly encapsulating cytoskeletal proteins in GUVs with high encapsulation efficiency. In this method, first, lipid-monolayer droplets are generated by emulsifying a protein solution of interest in a lipid/oil mixture. After being added into a rotating 3D-printed chamber, these lipid-monolayered droplets then pass through a second lipid monolayer at a water/oil interface inside the chamber to form GUVs that contain the protein system. This method simplifies the overall procedure of encapsulation within GUVs and speeds up the process, and thus allows us to confine and observe the dynamic evolution of network assembly inside lipid bilayer vesicles. This platform is handy for studying the mechanics of cytoskeleton-membrane interactions in confinement.
Keywords: Emulsion transfer, cDICE, GUVs, bottom-up reconstitution, actin, fascin
SUMMARY:
This article introduces a simple method for expeditious production of giant unilamellar vesicles with encapsulated cytoskeletal proteins. The method proves to be useful for bottom-up reconstitution of cytoskeletal structures in confinement and cytoskeleton-membrane interactions.
INTRODUCTION:
Lipid bilayer compartments are used as model synthetic cells for studying enclosed organic reactions and membrane-based processes or as carrier modules in drug delivery applications1, 2. Bottom-up biology with purified components requires minimal experimental systems to explore properties and interactions between biomolecules, such as proteins and lipids3, 4. However, with the advancement of the field, there is an increased need for more complex experimental systems that better imitate the conditions in biological cells. Encapsulation in GUVs is a practical approach that can offer some of these cell-like properties by providing a deformable and selectively permeable lipid bilayer and a confined reaction space. In particular, in vitro reconstitution of cytoskeletal systems, as models of synthetic cells, can benefit from encapsulation in membrane compartments5. Many cytoskeletal proteins bind and interact with the cell membrane. As most cytoskeletal assemblies form structures that span the entirety of the cell, their shape is naturally determined by cell-sized confinement6.
Different methods are used to generate GUVs, such as the swelling7, 8, small vesicle fusion9, 10, emulsion transfer11, 12, pulsed jetting13, and other microfluidic approaches14–18. Although these methods are still utilized, each has its limitations. Thus, a robust and straightforward approach with a high yield of GUV encapsulation is highly desirable. Although techniques such as spontaneous swelling and electroswelling are widely adopted for the formation of GUVs, these methods are primarily compatible with specific lipid compositions19, low salt concentration buffers20, smaller encapsulant molecular size21, and require a high volume of the encapsulant. Fusing multiple small vesicles into a GUV is inherently energetically unfavorable, thus requiring specificity in charged lipid compositions9 and/or external fusion-inducing agents, such as peptides22 or other chemicals. Emulsion transfer and microfluidic methods, on the other hand, may require droplet stabilization through surfactant and solvent removal after bilayer formation, respectively21, 23. The complexity of experimental setup and device in microfluidic techniques such as pulsed jetting impose an additional challenge24. cDICE is an emulsion-based method derived from similar principles governing emulsion transfer25, 26. An aqueous solution (outer solution) and a lipid-oil mixture are stratified by centrifugal forces in a rotating cylindrical chamber (cDICE chamber) forming a lipid saturated interface. Shuttling lipid monolayered aqueous droplets into the rotating cDICE chamber results in zipping of a bilayer as droplets cross the lipid-saturated interface into the outer aqueous solution25, 27. The cDICE approach is a robust technique for GUV encapsulation. With the presented modified method, not only the high vesicle yield typical for cDICE with a significantly shorter encapsulation time (a few seconds) is achieved but GUV generation time that allow for the observation of time-dependent processes (e.g., actin cytoskeletal network formation) is significantly reduced. The protocol takes about 15–20 min from the start to GUV collection and imaging. Here, GUV generation is described using the cDICE method for encapsulating actin and actin-binding proteins (ABPs). However, the presented technique is applicable for encapsulating a wide range of biological reactions and membrane interactions, from the assembly of biopolymers to cell-free protein expression to membrane fusion-based cargo transfer.
PROTOCOL:
1. Preparation of oil-lipid-mixture
NOTE: The step needs to be performed in a fume hood following all the safety guidelines for handling chloroform.
1.1. Take 0.5 mL of chloroform in a 15 mL glass vial. Add 88 μL of 25 mg/mL dioleoyl-phosphocholine (DOPC), 9.3 μL of 50 mg/mL cholesterol, and 5 μL of 1 mg/mL dioleoyl-phosphoethanolamine-lissamine rhodamine B (rhodamine PE) (see Table of Materials) into the 15 ml glass vial.
Table of Materials
| Name of Material/Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| 18:1 Liss Rhod PE lipid in chloroform | Avanti Polar Lipids | 810150C | |
| Actin from rabbit skeletal muscle | Cytoskeleton | AKL99-A | |
| ATTO 488-actin from rabbit skeletal muscle | Hypermol | 8153-01 | |
| Axygen microtubes (200 μL) | Fisher Scientific | 14-222-262 | for handling ABPs |
| Black resin | Formlabs | RS-F2-GPBK-04 | |
| Cholesterol (powder) | Avanti Polar Lipids | 700100P | |
| Choloroform | Sigma Aldrich | 67-66-3 | |
| Clear resin | Formlabs | RS-F2-GPCL-04 | |
| Density gradient medium (Optiprep) | Sigma-Aldrich | D1556 | |
| DOPC lipid in chloroform | Avanti Polar Lipids | 850375C | |
| Fascin | homemade | N/A | |
| F-buffer | homemade | N/A | |
| Fisherbrand microtubes (1.5 mL) | Fisher Scientific | 05-408-129 | |
| FS02 Sonicator | Fischer Scientific | FS20 | |
| G-buffer | homemade | N/A | |
| Glucose | Sigma-Aldrich | 158968 | |
| Mineral oil | Acros Organics | 8042-47-5 | |
| Silicone oil | Sigma-Aldrich | 317667 | |
| 96 Well Optical Btm Pit PolymerBase | ThermoFisher Scientific | 165305 | |
| Olympus IX81 Inverted Microscope | Olympus | IX21 | |
| CSU-X1 Confocal Scanner Unit | YOKOGAWA | CSU-X1 | |
| iXon X3 camera | Andor | DU-897E-CS0 | |
| Olympus PlanApo N 60x Oil Microscope Objective | Olumpus | 1-U2B933 |
NOTE: The final mole fractions of DOPC and cholesterol in silicone oil/mineral oil are 69.9% and 30%, respectively. It was established that 20–30 mol% cholesterol is an optimized concentration for membrane fluidity and stability of GUVs generated using the presented technique28, 29. Physiologically, these values are well within the cholesterol concentration range found in mammalian cell plasma membranes6. Lipid stocks are acquired as solutions in chloroform and stored at −20 °C. Lipid stock vials should be acclimated to room temperature before they are opened.
1.2. Pipette 7.2 mL of silicone oil and 1.8 mL of mineral oil in a second 15 mL vial (see Table of Materials). Generally, this needs to be done in a low-humidity glove box, mainly if the mineral oil is reused.
1.3. Mix the oils by vortexing at the maximum rotational speed (3200 RPM) for 10 s, add the mixture to the vial containing the lipid-in-chloroform mixture and immediately place it on the vortex mixer. Vortex for 10–15 s at the maximum rotational speed (3200 RPM). The resulting lipids-in-oil mix should be slightly cloudy, as the lipids are not fully dissolved in the oil but rather dispersed as small aggregates27.
1.4. Put the lipid-in-oil dispersion in a bath sonicator (see Table of Materials) with ultrasonic power of 80 W and operating frequency of 40 kHz at room temperature for 30 min. Use the mixture immediately or store at 4 °C for a maximum of 24 hours.
2. Vesicle generation
2.1. Mount the 3D-printed shaft (Supplementary File 1) made from black resin (see Table of Materials) on the benchtop stir plate and set rotational speed to 1200 RPM.
2.2. Mount the 3D-printed cDICE chamber (Supplementary File 2) made from clear resin (see Table of Materials) on the shaft (Figure 1A–B).
Figure 1: Experimental setup for generating GUVs.
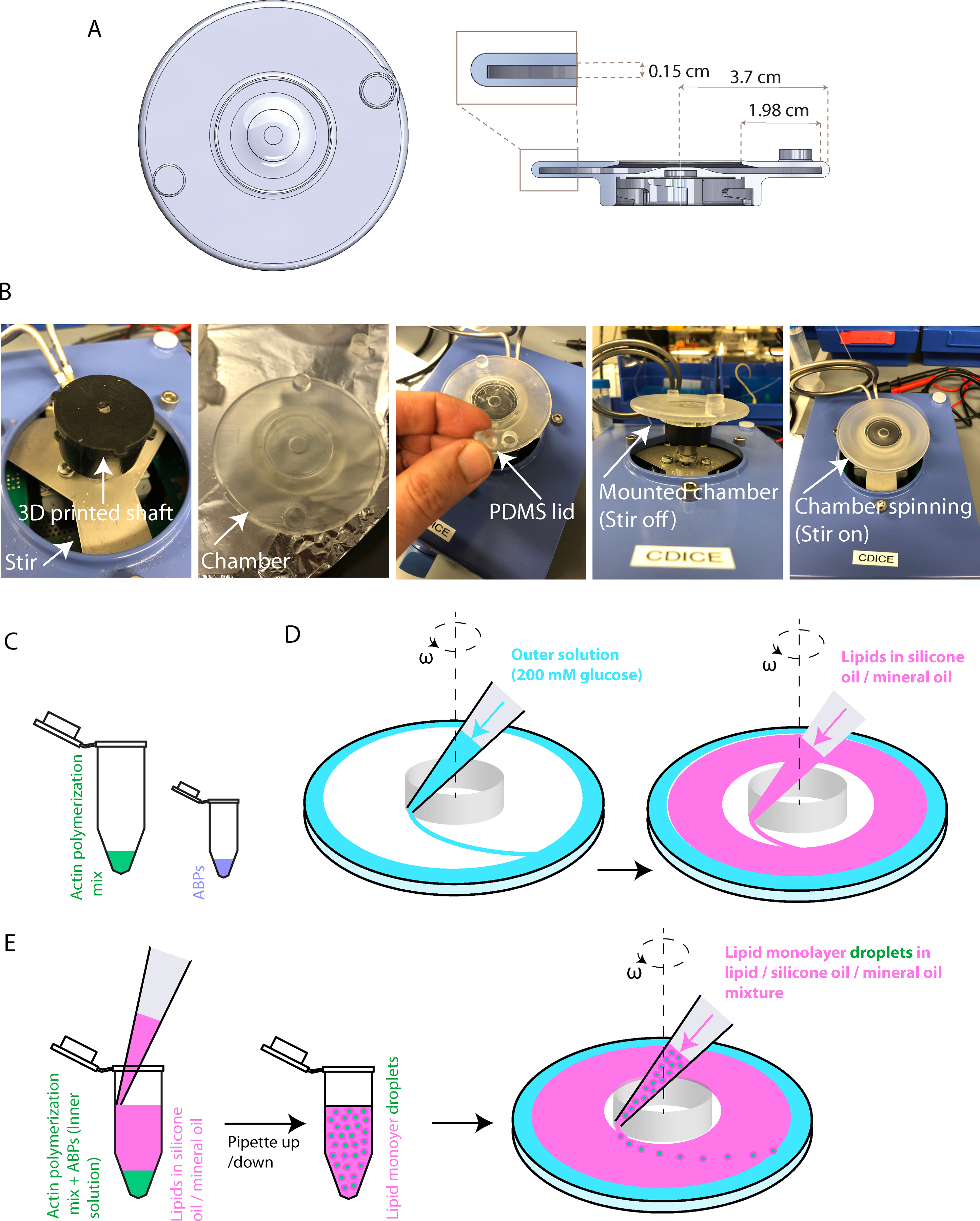
(A) Top view and side section view of the cDICE chamber. (B) Photos of the setup for the spinning chamber. (C-E) Schematic illustrations of stepwise procedures for generation of GUVs.
2.3. Prepare actin and actin-binding proteins (ABP) solutions separately in a total volume of 20 μL.
2.3.1. Prepare 1–10 μM of actin in globular actin buffer (G-buffer), including 10% ATTO 488 actin (see Table of Materials).
NOTE: 1x G-buffer comprises 5 mM of Tris-HCl, pH 8.0, and 0.2 mM of CaCl2.
2.3.2. Add filamentous actin polymerization buffer (F-buffer) to begin actin polymerization on ice. Keep the solutions on ice to slow down actin polymerization before the addition of a crosslinker.
NOTE: The 1x F-buffer contains 50 mM of KCl, 2 mM of MgCl2, and 3 mM of ATP in 100 mM of Tris, pH 7.5.
2.3.3. Wait for 15 min to allow for initiation of actin polymerization on ice before adding crosslinkers of interest at the desired molar ratio. Keep the solution on ice until encapsulation.
2.3.4. Prepare ABPs separately in a microtube (Figure 1C).
Note: This step is performed when a combination of ABPs (e.g., myosin, α-actinin, fascin, Arp2/3 complex) are to be encapsulated with actin28–30. In such cases, the desired amount of each ABP is drawn from its stock and added into the microtube designted for ABPs. As the total inner solution is to be 20 μL, stock aliquots of ABPs must be prepared in such a way that the desired amount of the total ABP mixture does not exceed 5–6 μL. The only ABP used in the representative results here is 1.57 μL fascin from a 1.75 mg/mL stock fascin, and it can be direcly added into the actin solution in step 2.7. This corresponds to 2.5 μM fascin in the inner solution.
2.4. Add 7.5% of density gradient medium (see Table of Materials) into the actin solution to create a density gradient between the outer and the inner aqueous phase to facilitate GUV sedimentation.
2.5. Dispense 700 μL of the outer solution of 200 mM of glucose into the chamber (Figure 1D, left).
NOTE: The osmolarity of the inner solution determines the concentration of glucose. For these experiments, the osmolarity of the inner solution is ~200 mOsm, so a 200 mM glucose solution is used as the outer solution.
2.6. Add a sufficient amount of lipid-oil mixture (>3 mL based on the chamber size) into the chamber until 60%−80% of the chamber is filled (Figure 1D, right). An interface will be formed between the lipid-oil mixture and the outer solution.
2.7. Transfer ABPs into the actin solution and, using a regular 100–1000 μL pipette, immediately transfer the 700 μL of the lipid-oil mixture into the actin-ABP mixture (Figure 1E, left). Pipette up and down 8 times to generate cell-sized lipid-monolayer droplets with diameter in the range of 7–100 μm (Figure 1E, middle).
NOTE: Step 2.7 needs to be completed in a few seconds to avoid any actin network assembly before encapsulation. Thus, before performing the step, ensure that pipette tips are already inserted into the pipettes and ready to transfer the mixtures.
2.8. Using the same 100–1000 μL pipette, immediately dispense the entire emulsion into the rotating chamber. Droplets will acquire a second leaflet of lipids by crossing the lipid monolayer at the oil-outer solution interface, thereby forming GUVs (Figure 1E, right).
2.9. Remove the chamber from the stir plate and discard most of the lipid-oil mixture by tilting the chamber in the waste container so that a large portion of the lipid-oil mixture is drained from the large opening at the center of the chamber.
NOTE: This way, the lipid-oil mixture is drawn off the chamber, avoiding mixing of lipid-oil mixture with the outer solution in the next step.
2.10. Hold the chamber with its lid facing towards the user. Open the chamber lid and slightly tilt the chamber towards the user. The interface between the outer solution containing GUVs and the lipid-oil mixture is visible from the chamber opening (where the lid is located).
2.11. Using a pipette, collect enough outer solution containing GUVs and dispense 50–300 μL of the outer solution into a 96-well plate to obtain an appropriate density of GUVs.
NOTE: Following this protocol, a total of about 2 × 105 GUVs are released in the outer solution inside the chamber. The GUV dispersity was not quantified; however, the diameter of about 90% of GUVs is in the range of 12–25 μm. GUVs with any diameter in the range of 7–50 μm can be found in the population. Depending on the encapsulated density gradient medium, GUV size, and solution depth in the well plate, it takes 2–15 min for GUVs to settle down on the surface. The yield of GUVs with reconstituted actin bundles is about 90%.
3. Imaging and 3D image reconstruction
3.1. Set up the 96 well plate on the stage of an inverted microscope equipped with a spinning disk (or laser scanning) confocal unit, an EMCCD or an sCMOS camera, and an oil immersion 60x objective lens (see Table of Materials).
3.2. Focus on any region of interest (ROI) and take a z-stack image sequence from the ROI with a z-step interval of 0.5 μm.
NOTE: Because GUVs can be slightly displaced on the surface over time, it is recommended to capture a multi-wavelength set of images at each z-plane if multiple fluorophores are being imaged, i.e., take a set of 561 nm and 488 nm images at each z-lane at a time to capture ATTO 488 actin and Rhod PE images.
3.3. Save each z-stack image sequence in .tiff format.
3.4. Open an image sequence of interest in an image processing software (ImageJ/Fiji). Identify the image with the highest intensity. Hold “ctrl+shift+c” to open the Brightness & Contrast window and click on Reset.
3.5. From the ImageJ/Fiji menu, go to Analyze > Set Scale and enter the known physical distance and its unit for each image pixel.
3.6. From the ImageJ/Fiji menu, go to Image > Stacks > 3D project to reconstruct a 3D image from the z-stack. Set “Projection method” as Brightness Point, “Slice Spacing (μm)” as 0.5, and checkmark Interpolate. The default options can be used for the rest of the settings.
NOTE: z-intervals in some microscopes might not be calibrated, and the actual movement in z-direction might slightly differ from the entered z-interval (i.e., 0.5 μm). In such cases, a 3D calibration specimen such as fluorescent microspheres with a known diameter can be used to obtain the actual z-interval. This value will thus be used as “Slice Spacing (μm)” for 3D projection.
REPRESENTATIVE RESULTS:
To demonstrate the successful generation of cytoskeletal GUVs using the current protocol, fascin-actin bundle structures in GUVs were reconstituted. Fascin is a short crosslinker of actin filaments which forms stiff parallel-aligned actin bundles and is purified from E. coli as Glutathione-S-Transferase (GST) fusion protein29. 5 μM of actin was first reconstituted, including 0.53 μM of ATTO488 actin in the actin polymerization buffer and 7.5% of the density gradient medium. Upon adding fascin at a concentration of 2.5 μM and encapsulation of the fascin-actin mixture, actin bundle structures were formed in GUVs. z-stack confocal image sequences of the encapsulated actin bundle structures in the Rhod PE-labeled GUVs were captured 1 h post-encapsulation (Figure 2A). Using this protocol, inherent competition and sorting of the encapsulated actin crosslinkers, α-actinin, and fascin, which, together, form different actin bundle patterns in a GUV-size dependent manner, was previously demonstrated29.
Figure 2: Encapsulation of actin bundle structures.
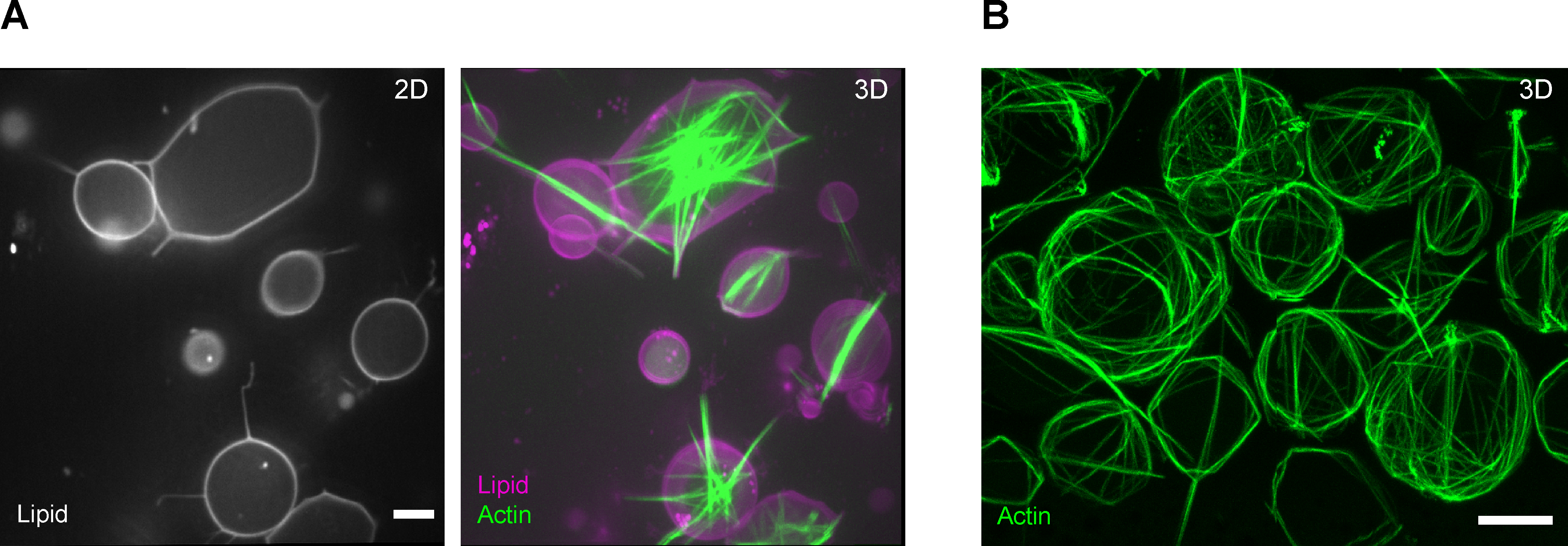
(A) The images show representative fluorescence confocal slices of GUVs (left) and maximum projections of a confocal z-stack of actin and lipid channels (right). Fascin, 2.5 μM; actin, 5 μM (including 10% ATTO 488 actin). Scale bar = 10 μm. (B) Encapsulation of actin bundle structures using conventional cDICE. The image shows a representative maximum projection of confocal fluorescence images of encapsulated actin bundles formed in the presence of fascin. Fascin, 0.9 μM; Actin, 6 μM. Scale bar = 10 μm.
Like the modified inverted emulsion approach presented here, the traditional cDICE process generates cytoskeletal GUVs with high yield yet requires a syringe pump and tubing setup for controlled injection of protein solution into the rotating chamber at low flow rates the order of nanoliters per second25, 31. In this approach, the emulsion is directly generated in the rotating cDICE chamber; a thin capillary is inserted in the oil phase. The protein solution is injected through a syringe pump. Droplets form and are sheared off at the capillary tip before they travel towards the aqueous outer phase, where they turn into GUVs, similar to the method described above. Figure 2B shows vesicles that encapsulate a reaction mixture using this approach. The reaction mix contains 6 μM of actin which is bundled by 0.9 μM of fascin. Here, the two methods and their results are not being compared but note that they both generate a high yield of GUVs.
DISCUSSION:
Different methods of generating GUVs have been explored for the creation of synthetic cells. However, the complexity of the procedures, extended time to attain encapsulation, restriction of lipid types and molecular composition of the encapsulant, need for non-physiological chemicals to facilitate encapsulation, low GUV yield, and inconsistencies in encapsulation efficiency have continued to challenge researchers in this field. Considering the wide range of potential studies that can be embarked in bottom-up synthetic biology, a seamless high throughput GUV encapsulation approach that is compatible with different lipid compositions and can encapsulate any molecules regardless of size may spur new opportunities to study complex biomimicking synthetic systems. The cDICE method has eliminated most challenges and limitations inherent to prior GUV generation methods.
The approach and governing principles to generate GUVs using the cDICE method predates the platform and have been implemented in earlier techniques such as the inverted emulsion transfer12. However, the inverted emulsion transfer method has limitations such as low vesicle yield and heterogeneity of vesicles. For the cDICE method presented here, lipids are dispersed in oil in the form of aggregates of tens of nanometers (size of lipid aggregate is dependent upon the overall concentration of lipids)27. Dispersion of lipids is in two miscible oils, where one (mineral oil) can dissolve lipids and a second oil (silicon oil) that is not miscible with lipids. This creates lipid aggregate coacervates by way of solvent-shifting32. This particular dispersion approach facilitates instant monolayer saturation of aqueous droplets, faster renewal of lipids at the oil-aqueous interface as aqueous droplets continuously cross the lipid-saturated oil-aqueous interface. This also subsequently improves bilayer zipping to form GUVs and increases GUV throughput. The centrifugal forces generated by the rotating chamber are optimal for shuttling polydispersed droplets across the lipid-saturated interface. The original version of the cDICE method utilizes a microcapillary nozzle to inject the inner solution into the oil-lipid mixture. In this approach, shearing forces created by the rotating oil-lipid mixture generate aqueous droplets, eventually transforming into GUVs as described. However, with the intent to reduce the time taken to prepare injection platform, especially critical for fast reactions, such as actin network assembly and potential clogging of the microcapillary, aqueous droplets with lipid monolayers are now generated by adding the inner solution directly to the oil-lipid mixture and pipetting up and down. This approach eliminates the time lag in GUV encapsulation for a fast reaction experiment.
Amongst the challenges caused by earlier GUV generation methods is the restriction of lipid types (charge of lipid and phase of lipid) depending on the technique of GUV generation. Multiple lipid types, including DOPC, dioleoyl-glycero-hosphoserine (DOPS), DGS (dioleoyl-glycero-succinate), dimyristoyl-glycero-phosphocholine (DMPC), and combinations of different lipids and cholesterol at varying concentrations were tested. For all conditions, the cDICE method is shown to form GUVs with a high encapsulation efficiency at a consistently high GUV yield. Furthermore, the cDICE method has also been shown to effectively encapsulate different cellular components, including cytoskeletal proteins, cell-free expression reactions, crowding agents, dyes, and other cellular molecules of different sizes without a loss of encapsulation efficiency or decreased throughput. Furthermore, like some microfluidic GUV generation methods33, the modified cDICE can potentially permit the generation of asymmetric GUVs for future work. Different lipid compositions can be used for the inner leaflet and the outer leaflet since monolayer droplets are formed separately (inside a microtube by pipetting up and down) before zipping the bilayer inside the cDICE chamber. It was also confirmed that the lipid-oil mixture could last up to 3 weeks if kept in tight seal glass vials at 4 °C while maintaining high yield and high GUV encapsulation efficiency. Sedimentation of lipids is observed when the lipid-oil mixture is kept for long; however, one can simply vortex the lipid-oil mixture before encapsulation to re-disperse lipid aggregates. It is important to note that encapsulation quality can become compromised when the lipid-oil mixtures are kept for a longer time, as indicated by more significant than usual lipid aggregates in the lipid-oil mix. Although not tested, these aggregates could potentially result in imperfection in bilayer zipping, and the aggregates may also end up being encapsulated with inner solution compromising the desired chemical environment.
The limitation of the presented modified approach to form aqueous droplets is in generating uniformity in the size of droplets. Although this can be improved by using microcapillary injection of inner solution at different flow rates to regulate GUV sizes, it is less desirable if one wants to monintor fast reactions like actin assembly in encapsulated GUVs. By making droplets by pipetting up and down that results in different GUV sizes, one can analyze populations of similar size vesicles. Concerns about possible oil retention in bilayers impede the adoption of most GUV generation techniques, including emulsion-based GUV generation techniques such as cDICE24. However, the amount of oil remaining in the membrane may be reduced by using organic agents such as 1-octanol, which can be removed after generating the vesicles34, 35. Future modifications to the method, possibly by changing solvent composition, need to be investigated.
There are many areas in bottom-up synthetic biology that are yet to be investigated and perhaps require cell-mimicking confinements of GUVs. Such experimental endeavors necessitate GUV generation platforms like cDICE to generate GUVs robustly while efficiently encapsulating various molecules of interest. Many cellular processes occur faster than the time it takes to encapsulate molecules using prior GUV generation techniques. As described here, actin solutions are encapsulated quickly enough to observe vesicle deformation resulting from actin network assembly. Such synthetic cells with reconstituted actin cytoskeleton have revealed features of actin network organization in the presence of different crosslinkers5, 28, 36 and membrane remodeling28, 30, 31. They will inspire future work to create more sophisticated synthetic cells.
Supplementary Material
Supplementary File 1: 3D printed shaft design.
Supplementary File 2: Design for the 3D-printed cDICE chamber.
ACKNOWLEDGMENTS:
APL acknowledges support by a Humboldt Research Fellowship for Experienced Researchers and from the National Science Foundation (1939310 and 1817909) and National Institutes of Health (R01 EB030031).
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/63332.
DISCLOSURES:
The authors declare no conflicts of interest.
REFERENCES:
- 1.Groaz A et al. Engineering spatiotemporal organization and dynamics in synthetic cells. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 13 (3), e1685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diltemiz SE et al. Use of artificial cells as drug carriers. This journal is Cite this: Mater. Chem. Front. 5, 6672, doi: 10.1039/d1qm00717c (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu AP, Fletcher DA Biology under construction: in vitro reconstitution of cellular function. Nature Reviews Molecular Cell Biology 2009 10:9. 10 (9), 644–650, doi: 10.1038/nrm2746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganzinger KA, Schwille P More from less – bottom-up reconstitution of cell biology. Journal of Cell Science. 132 (4), doi: 10.1242/JCS.227488 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Bashirzadeh Y, Liu AP Encapsulation of the cytoskeleton: towards mimicking the mechanics of a cell. Soft Matter. 15 (42), 8425–8436 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J Actin Dynamics, Architecture, and Mechanics in Cell Motility. 10.1152/physrev.00018.2013. 94 (1), 235–263, doi: (2014). [DOI] [PubMed] [Google Scholar]
- 7.Angelova MI, Dimitrov DS Liposome electroformation. Faraday discussions of the Chemical Society. 81, 303–311 (1986). [Google Scholar]
- 8.Tsumoto K, Matsuo H, Tomita M, Yoshimura T Efficient formation of giant liposomes through the gentle hydration of phosphatidylcholine films doped with sugar. Colloids and Surfaces B: Biointerfaces. 68 (1), 98–105 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Bailey AL, Cullis PR Membrane fusion with cationic liposomes: effects of target membrane lipid composition. Biochemistry. 36 (7), 1628–1634 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Haluska CK, Riske KA, Marchi-Artzner V, Lehn J-M, Lipowsky R, Dimova R Time scales of membrane fusion revealed by direct imaging of vesicle fusion with high temporal resolution. Proceedings of the National Academy of Sciences. 103 (43), 15841–15846 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura K, Suzuki H, Toyota T, Yomo T Size control of giant unilamellar vesicles prepared from inverted emulsion droplets. Journal of colloid and interface science. 376 (1), 119–125 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Pautot S, Frisken BJ, Weitz DA Production of unilamellar vesicles using an inverted emulsion. Langmuir. 19 (7), 2870–2879 (2003). [Google Scholar]
- 13.Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proceedings of the national academy of sciences. 105 (12), 4697–4702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett MK, Calakos N, Scheller RH Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 257 (5067), 255–259, doi: 10.1126/SCIENCE.1321498 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Lee YL, Burke B LINC complexes and nuclear positioning. Seminars in Cell and Developmental Biology. 82, 67–76, doi: 10.1016/j.semcdb.2017.11.008 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Noireaux V, Liu AP The New Age of Cell-Free Biology. doi: 10.1146/annurev-bioeng-092019 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Berhanu S, Ueda T, Kuruma Y Artificial photosynthetic cell producing energy for protein synthesis. Nature Communications 2019 10:1. 10 (1), 1–10, doi: 10.1038/s41467-019-09147-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho KKY, Murray VL, Liu AP Engineering artificial cells by combining HeLa-based cell-free expression and ultrathin double emulsion template. Methods in Cell Biology. 128, 303–318, doi: 10.1016/BS.MCB.2015.01.014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akashi K, Miyata H, Itoh H, Kinosita K Jr Preparation of giant liposomes in physiological conditions and their characterization under an optical microscope. Biophysical journal. 71 (6), 3242–3250 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méléard P, Bagatolli LA, Pott T Giant unilamellar vesicle electroformation: From lipid mixtures to native membranes under physiological conditions. Methods in enzymology. 465, 161–176 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Walde P, Cosentino K, Engel H, Stano P Giant Vesicles: Preparations and Applications. ChemBioChem. 11 (7), 848–865, doi: 10.1002/cbic.201000010 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Pécheur E-I, Martin I, Ruysschaert J-M, Bienvenüe A, Hoekstra D Membrane fusion induced by 11-mer anionic and cationic peptides: a structure− function study. Biochemistry. 37 (8), 2361–2371 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Morigaki K, Walde P Giant vesicle formation from oleic acid/sodium oleate on glass surfaces induced by adsorbed hydrocarbon molecules. Langmuir. 18 (26), 10509–10511 (2002). [Google Scholar]
- 24.Majumder S, Wubshet N, Liu AP Encapsulation of complex solutions using droplet microfluidics towards the synthesis of artificial cells. Journal of Micromechanics and Microengineering. 29 (8), 83001 (2019). [Google Scholar]
- 25.Abkarian M, Loiseau E, Massiera G Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter. 7 (10), 4610–4614 (2011). [Google Scholar]
- 26.Cauter L Van de et al. Optimized cDICE for Efficient Reconstitution of Biological Systems in Giant Unilamellar Vesicles. ACS Synthetic Biology. 10 (7), 1690–1702, doi: 10.1021/ACSSYNBIO.1C00068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claudet C, In M, Massiera G Method to disperse lipids as aggregates in oil for bilayers production. The European Physical Journal E. 39 (1), 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Bashirzadeh Y, Wubshet NH, Liu AP Confinement Geometry Tunes Fascin-Actin Bundle Structures and Consequently the Shape of a Lipid Bilayer Vesicle. Frontiers in molecular biosciences. 7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bashirzadeh Y et al. Actin crosslinker competition and sorting drive emergent GUV size-dependent actin network architecture. Communications Biology. 4 (1), doi: 10.1038/s42003-021-02653-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashirzadeh Y, Moghimianavval H, Liu AP Encapsulated actomyosin patterns drive cell-like membrane shape changes. bioRxiv. 2021.10.20.465228, doi: 10.1101/2021.10.20.465228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litschel T et al. Reconstitution of contractile actomyosin rings in vesicles. Nature communications. 12 (1), 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitale SA, Katz JL Liquid droplet dispersions formed by homogeneous liquid− liquid nucleation:“The Ouzo effect.” Langmuir. 19 (10), 4105–4110 (2003). [Google Scholar]
- 33.Pautot S, Frisken BJ, Weitz DA Engineering asymmetric vesicles. 100 (19), at <www.pnas.orgcgidoi10.1073pnas.1931005100> (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deshpande S, Dekker C On-chip microfluidic production of cell-sized liposomes. Nature Protocols 2018 13:5. 13 (5), 856–874, doi: 10.1038/nprot.2017.160 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Deshpande S, Caspi Y, Meijering AEC, Dekker C Octanol-assisted liposome assembly on chip. Nature Communications. 7, doi: 10.1038/ncomms10447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wubshet NH, Bashirzadeh Y, Liu AP Fascin-induced actin protrusions are suppressed by dendritic networks in GUVs. Molecular Biology of the Cell. mbc-E21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: 3D printed shaft design.
Supplementary File 2: Design for the 3D-printed cDICE chamber.


