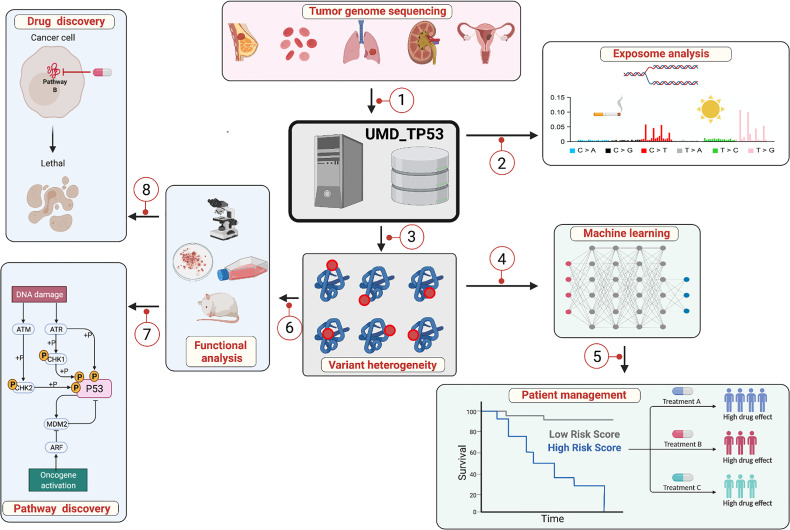
Figure 1.
The locus-specific database UMD_TP53: a central hub for multifactorial analysis. 1: TP53 variants and patient information are collected and stored in a relational database specifically developed for the storage and the analysis of genetic variants. 2: Exposome analysis: influence of the external and internal environment on the landscape of mutational events to identify the links between exposure to various types of carcinogens, specific mutational events in the TP53 gene and the development of specific cancers. 3: More than 7,000 different TP53 variants have been discovered in various types of cancer with heterogeneous LOF and GOF. 4: Multiple bioinformatics tools, including machine learning, have been developed to predict and classify TP53 variants. 5: Genome-based prognostic biomarkers can be used for several cancer types for potential incorporation into clinical prognostic staging systems or practice guidelines such as TP53 and CLL. 6: Analysis of TP53 variants points to the various functional domains of the protein essential for tumor suppression. 7: Functional analysis has led to the identification of the multiple pathways regulated by TP53. 8: Small molecules have been developed that specifically target missense TP53 variants and restore p53 transcriptional activity, thereby enabling tumor regression. Although this figure describes the TP53 database, the various aspects can be applied to other genes as well.