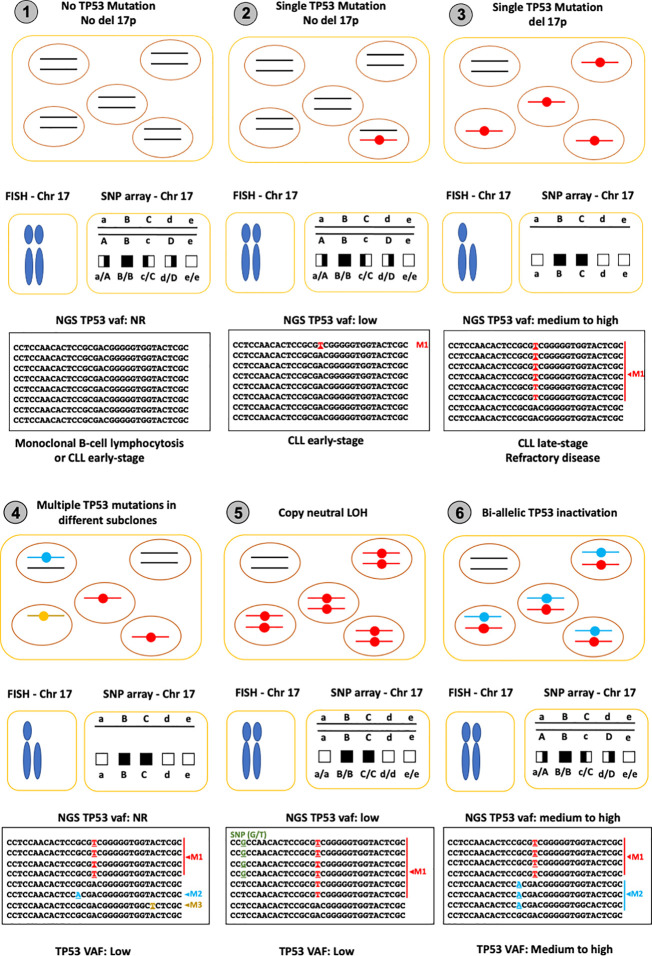
Figure 4.
TP53 status in CLL patients, a snapshot. The top panel displays a schematic view of the tumor with the two TP53 alleles. The middle panel shows cytogenetic analysis performed by FISH (left) or by SNP arrays (right). The lower panel displays an example of the read alignments from NGS. 1: No TP53 mutation: In monoclonal B-cell lymphocytosis, TP53 mutation and 17p deletion are very rare, leading to negative results for both FISH and genetic analysis. 2: TP53 mutation without LOH: In early stages of CLL, the frequency of TP53 mutation is low (less than 10%) with many cases showing no LOH. Sensitive sequencing analysis with NGS is able to identify low VAF TP53 variants (variant M1 in the lower panel). 3: TP53 mutation with LOH: In late-stage or relapsing disease, TP53 mutations associated with 17p deletion can be found in 30 to 50% of CLL patients. In the majority of cases, VAF is greater than 50% due to the loss of the second allele. This situation is commonly seen in CLL. 4: Multiple TP53 and LOH: in both early and late-stage disease, FASAY (functional analysis of separated alleles of p53 on yeast) or SMRT (single-molecule, real-time sequencing) has demonstrated a high level of intratumoral heterogeneity in CLL with the presence of multiple independent subclones expressing different pathogenic TP53 variants (M1, M2 and M3 in the lower panel). Although 17p deletion is often observed in these patients, it is difficult to determine if subclones expressing different TP53 variants are associated with it, and even more so if the VAF of the variant is low. 5: Copy neutral LOH: Following the initial mutational inactivation of one allele, the remaining wild-type allele is deleted concurrently with the duplication of the mutated allele, leading to copy neutral LOH (cnLOH). Detecting cnLOH is difficult and thus the frequency of the event is currently unknown. Without SNP array analysis and if the VAF of the variant is lower than 50%, this situation can be misidentified as a tumor without LOH. Tumors with VAF greater than 50% without obvious 17p deletion should be checked for cnLOH. 6: Bi-allelic mutations: Inactivation of the TP53 gene via different mutations in the two alleles is possible but difficult to distinguish from intratumoral heterogeneity. Although this situation is often described as plausible in many reviews, it has never been formally identified, as only single-cell sequencing would be able to validate bi-allelic TP53 inactivation.