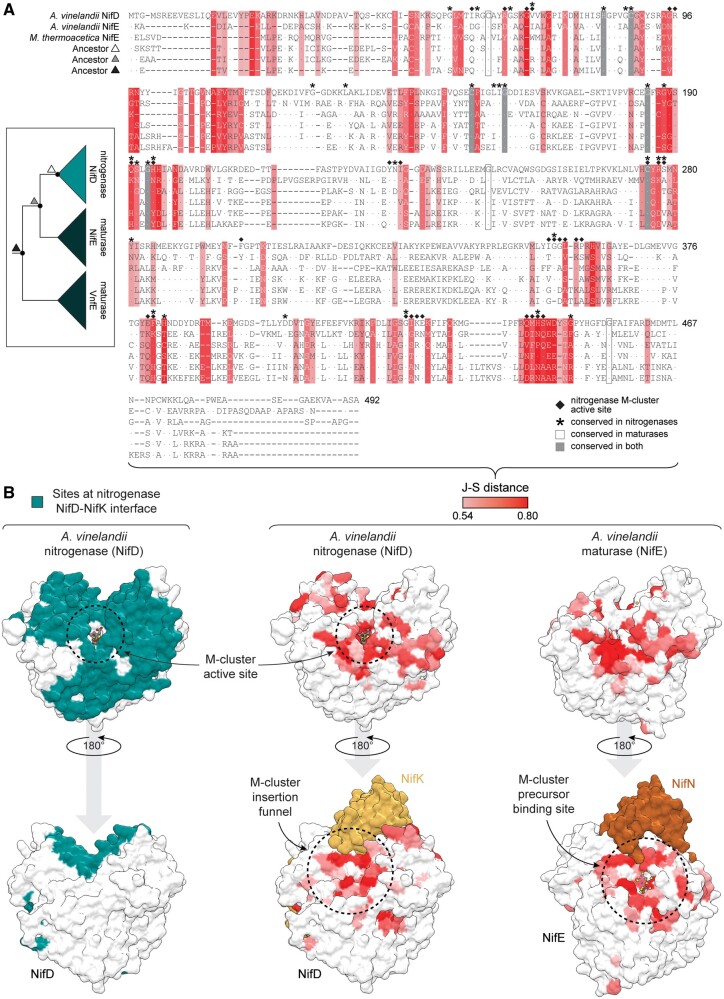
Fig. 2.
Structure and sequence maps of putative functionally divergent protein sites between nitrogenases and maturases. (A, B) Putative functionally divergent sites are defined as those above the 75th percentile J–S distance between nitrogenase NifD and maturase NifE proteins (see text for details). The J–S distance scale applies to both (A) and (B). (A) Maximum-likelihood ancestors (inferred from Tree-1; fig. 1C) aligned to representative extant nitrogenase (NifD) and maturase (NifE) sequences. Moorella thermoacetica NifE is an uncharacterized, putative maturase sequence. Ancestor triangle symbols correspond to labeled nodes in the simplified phylogeny (left) and match those in figure 3. Nitrogenase M-cluster active-site residues are defined as those within 5 Å of the M-cluster. Dots within the alignment indicate residue identity to Azotobacter vinelandii NifD. Site numbering based on A. vinelandii NifD. (B) Divergent sites mapped to aligned nitrogenase (center; A. vinelandii NifD, PDB 3U7Q) and maturase (right; A. vinelandii NifE, PDB 3PDI) subunit structures. Protein sites at the nitrogenase NifD-NifK interface (left) are defined as those within 5 Å of the NifK subunit.