Figure 2.
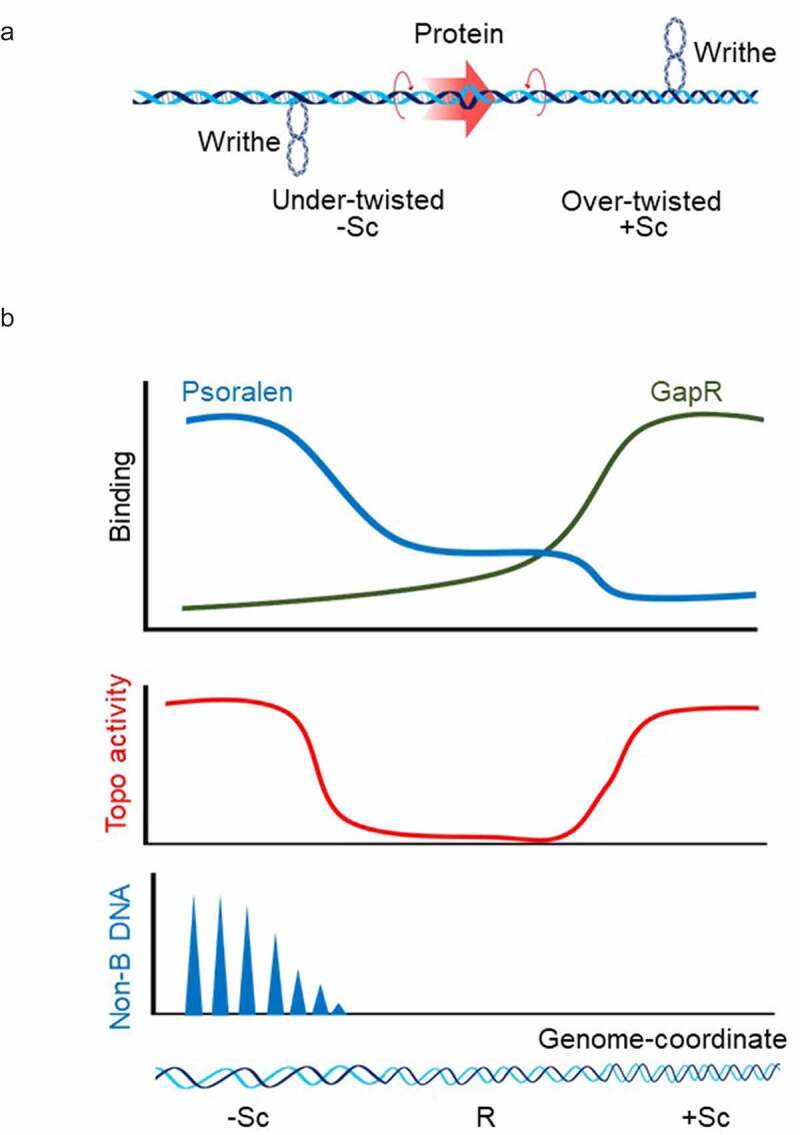
DNA Supercoiling in vivo (a) Twin-supercoiled domain model explaining the generation of dynamic positive supercoils ahead and negative supercoils behind of the protein complex that translocates along the DNA double helix [15]. Although this supercoiling regulates the variety of DNA transactions, excessive DNA supercoils will halt the further progression of translocating complex if not properly resolved. (b) Methods for detection of DNA supercoiling in vivo. Top panel: DNA supercoiling have been most frequently probed with psoralen. Psoralen freely crosses cellular membranes, intercalates between DNA bases and forms crosslinks between the two strands when exposed to UV light [127]. It has a different preference for relaxed, positively supercoiled, and negatively supercoiled DNA (blue curve). Taking advantage of this psoralen property, supercoiled DNA have been mapped in bacteria, yeast, Drosophila, and human cells [66,69,119,145]. Recently developed GapR-seq assay is based on the ability of the bacterial protein GapR to preferentially recognize overtwisted DNA (green curve). Chromatin immunoprecipitation of GapR combined with high-throughput sequencing was used to generate maps of positive supercoiling in bacteria and yeast [97]. Detection of topoisomerase activity sites (Middle panel) and non-B DNA structures (Bottom panel) are also powerful methods to predict DNA supercoiling in vivo [71,132,146]. There has been considerable concordance between the studies supporting the main prophecies of the twin-supercoiled domain model: negative torsional stress accumulated at the upstream promoter region of the active genes, while positive torsional stress accrues in a transcription-dependent manner in gene bodies and downstream to the 3’ ends of genes. – Sc (negatively supercoiled DNA); R (relaxed DNA); +Sc (positively supercoiled DNA). Blue triangle (Non-B DNA).
