Figure 1.
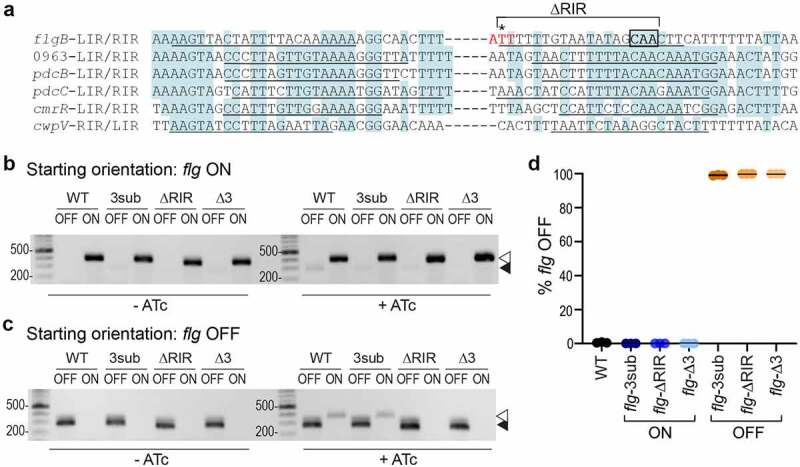
Mutations in flg RIR affect inversion in E. coli and C. difficile. a. Alignment of inverted repeats flanking the invertible DNA sequences affected by RecV. Shading denotes residues conserved in at least four of the six repeats. Putative inverted repeats are underlined. Hyphens between LIRs and RIRs represent the intervening sequences, which vary in length and nucleotide sequence. For the flgB-RIR, the site of cleavage by RecV is indicated with an asterisk. This nucleotide and the two adjacent residues, indicated in red, were deleted in flg-Δ3 constructs/strains. Note that the adenine 5’ of the cleavage site is present in the flg ON sequence, whereas a thymine is present in flg OFF; constructs for mutagenesis were created in both flg orientations. The conserved CAA nucleotides mutated in flg-3sub constructs/strains are boxed in black. Nucleotides deleted in flg-ΔRIR bacteria/constructs are indicated. LIR, RIR = left, right inverted repeats. Numbers indicate locus tags in C. difficile R20291. b, c. Orientation-specific PCR to examine flagellar switch inversion in E. coli bearing wildtype or mutated inverted repeat target sequences. The starting orientation of the flagellar switch is indicated: flg ON (b) or flg OFF (c). Absence or presence of ATc for induction of recV expression is shown (-ATc/+ATc). The flg ON and flg OFF products are indicated with white and black arrows, respectively. Shown are representative images of three independent experiments. (d) Analysis of flagellar switch orientation in C. difficile RIR mutants by quantitative orientation-specific PCR. Means and standard deviations for three biological replicates are shown.
