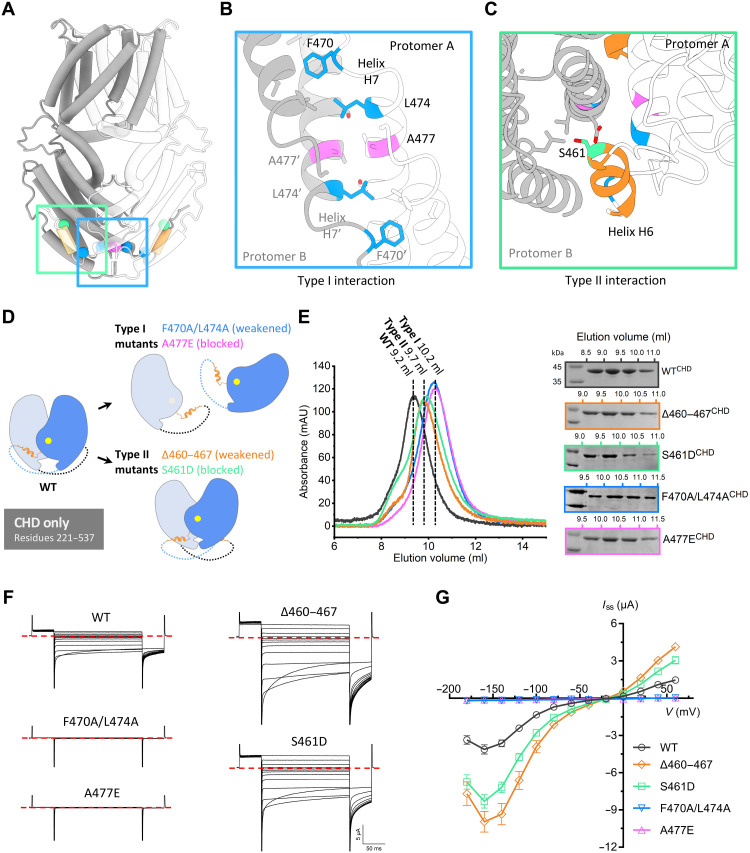
Fig. 7. Effect of dimeric interactions on channel activity.
(A) Cartoon of the GmALMT12/QUAC1 channel, with one protomer in gray and one in white. The domain-swapped helix H6 (residues 460 to 467) is colored in orange, and key residues at the dimeric interface are highlighted: S461 in green, A477 in magenta, and F470/L474 in blue. (B and C) Close-up views of the type I interaction (B) and type II interaction (C) at the cytoplasmic dimer interface. Residues at the dimeric interface are shown as sticks and colored as in (A). (D) Mutagenesis design for disrupting the interaction at the interface of dimeric CHDs. (E) Purification of the CHD proteins (residues 221 to 537, wild type or mutants) on a Superdex 75 (10/300) column. Compared to the wild type (peak at ~9.2 ml), the eluted peaks for the type I mutants (A477E and F470A/L474A) shift backward to ~10.2 ml, suggesting a disassociation of dimer into monomer. The elution peaks at ~9.7 ml for the type II mutants (∆460–467 and S461D) suggest altered conformations upon the removal of finger helix interaction. The eluted fractions were analyzed by SDS-PAGE, as indicated. (F and G) TEVC recording of GmALMT12/QUAC1 channel (wild type and mutants) in X. laevis oocytes. The same set of mutations, as in (D), was generated into full-length constructs for conductance measurement in the bath solution of 30 mM l-malate (data are means ± SEM, n ≥ 8). Representative current traces recorded at different voltages (from +60 to −180 mV in 20-mV decrement) are shown in (F). The steady-state current–voltage (Iss-V) relations are shown in (G). Data are means ± SEM, n ≥ 8.