Abstract
Chronic kidney disease (CKD) has become a widespread disease among people. It is related to various serious risks like cardiovascular disease, heightened risk, and end-stage renal disease, which can be feasibly avoidable by early detection and treatment of people in danger of this disease. The machine learning algorithm is a source of significant assistance for medical scientists to diagnose the disease accurately in its outset stage. Recently, Big Data platforms are integrated with machine learning algorithms to add value to healthcare. Therefore, this paper proposes hybrid machine learning techniques that include feature selection methods and machine learning classification algorithms based on big data platforms (Apache Spark) that were used to detect chronic kidney disease (CKD). The feature selection techniques, namely, Relief-F and chi-squared feature selection method, were applied to select the important features. Six machine learning classification algorithms were used in this research: decision tree (DT), logistic regression (LR), Naive Bayes (NB), Random Forest (RF), support vector machine (SVM), and Gradient-Boosted Trees (GBT Classifier) as ensemble learning algorithms. Four methods of evaluation, namely, accuracy, precision, recall, and F1-measure, were applied to validate the results. For each algorithm, the results of cross-validation and the testing results have been computed based on full features, the features selected by Relief-F, and the features selected by chi-squared feature selection method. The results showed that SVM, DT, and GBT Classifiers with the selected features had achieved the best performance at 100% accuracy. Overall, Relief-F's selected features are better than full features and the features selected by chi-square.
1. Introduction
The present era, especially the last two decades, can be named the era of big data where digital data is turning out to be very crucial more and more in various fields such as science, healthcare, technology, and society. Huge data volumes have been produced and generated from multiple sensor networks and mobile applications in almost all fields, including healthcare in specific, and this multitude of data volumes is what we call big data [1]. Wide variety of data sources such as streaming machines, high-end output instruments, visualizing, and knowledge extraction across these vast and diverse types of data pose a significant challenge when sufficient cutting-edge technologies and tools are not used. One of the most eminent technological challenges facing big data analytics lays in exploring ways that are adequate to obtain useful and relevant information for different user categories in an effective manner.
Nowadays, the different forms and types of data sources in healthcare are being gathered in both clinical and nonclinical environments, where the most crucial data in healthcare analytics is the digital copy of a patient's medical history. On that account, the process of designing and making up a distributed data system to handle big data is challenged by three main issues. The first challenge is that it is difficult to collect data from distributed locations because of the diverse and large data volume. The second challenge is that storage is the chief issue for heterogeneous and enormous datasets as big data system requires to store while allowing performance guarantee. The third challenge is more connected to big data analytics, specifically to enormous mining datasets in real time, and this includes visualization, prediction, and optimization [2].
Considering the difficulty imposed by these challenges, they require an up-to-date and advanced processing paradigm provided that the present data management systems do not provide adequate efficiency in handling the heterogeneous nature of data or the real-time aspect. Traditional database management systems cannot support the continuous increase in huge data size. To address these issues related to enormous and heterogeneous data storage, the research community has proposed a number of research works, such as Apache Spark, Apache Hadoop [3], Apache Kafka [4], and Apache Storm [5], to solve healthcare problems [6–8].
Chronic kidney disease (CKD) has received a lot of interest due to its high death rate. Chronic diseases have become a major hazard to emerging countries, according to the World Health Organization (WHO) [9]. CKD is a kidney illness that can be treated in its early stages, but it eventually leads to renal failure if not treated early. In 2016, chronic kidney disease claimed the lives of 753 million individuals globally, accounting for 336 million male deaths and 417 million female deaths [10]. Chronic renal disease can be prevented from progressing to kidney failure if diagnosed and treated early. Diagnosing chronic kidney disease early is the best method to treat it, while delaying treatment until it is too late may lead to renal failure, which necessitates dialysis or kidney transplantation to live normally. Therefore, global strategies for early detection and treatment of people with CKD are required. To mine hidden patterns from data for effective decision-making and to help doctors in making more accurate diagnoses, a computer-aided diagnosis system based on artificial intelligence strategies is needed for clinical information. Artificial intelligence techniques (machine learning and deep learning) have been used in the health field, namely, in disease prediction and diagnosis.
Chronic kidney disease (CKD) is a condition that affects the kidney's ability to function. In general, CKD is separated into phases, with renal failures occurring when the kidneys are no longer able to complete their roles of blood purification and mineral balance in the body [11]. According to the current estimates, CKD is more common in adults over 65 years old (38%) than in people aged 45–64 years (12%) and people aged 18–44 years (6%). Women have a rather higher rate of CKD (14%) than males [12].
Machine learning is an exciting field that focuses on studying huge amounts of data with multiple variables. Machine learning has basically developed from studying the theory of pattern recognition and computational learning in artificial intelligence; it presupposes computational methods, algorithms, and analysis techniques. From the perspective of Medical Sciences, machine learning undertakes to aid health specialists and doctors in carrying out scintillate and flawless diagnoses, choosing the best-fit medicines for patients, determining patients at high risk, and, most importantly, improving patients' physical condition with minimal cost.
Machine learning (ML) has demonstrated remarkable performance across a range of applications, such as speech recognition [13], computer vision [14], medical diagnostics [15], and engineering [16].
Being a constituent of the ML process, feature selection (FS) is a crucial preprocessing step that determines the most relevant attributes within a dataset. Removing unimportant and unnecessary attributes can result in less complicated and more accurate models. In this paper, two feature selection methods based on Apache Spark are used, namely, Relief-F [17] and chi-squared [18] feature selection method. Some of the research works have used ML techniques to predict CKD. For example, Charleonnan [19] et al. used four ML algorithms, K-nearest neighbors (KNN), support vector machine (SVM), logistic regression (LR), and decision tree (DT), to predict CKD. Other research works used hybrid ML algorithms that are integrated between feature selection methods and ML to predict CKD. Feature selection methods have been used to reduce the number of features and select the optimal subsets of features from the dataset. For example [20], authors used chi-square, correlation-based feature selection (CFS), and Lasso feature selection to select the essential features from the database. They applied artificial neural network (ANN), C5.0, LR, SVM, KNN, and RF to both full features and the selected features.
Recently, researchers have been using big data platforms such as Apache Spark [21] which is a large-scale data processing engine with a unified analytics engine. Spark is 100 times quicker than Hadoop in running workloads on large-scale clusters. It includes Java, Scala, Python, and R high-level APIs, as well as an efficient engine that supports broad execution graphs. It also includes a number of higher-level tools such as Spark SQL for SQL and structured data processing, MLlib, GraphX, and Structured Streaming.
Spark's machine learning (ML) [21] library is called MLlib. Its purpose is to make scalable and simple machine learning a reality. It provides, at a high level, tools such as classification, regression, clustering, and collaborative filtering as examples of machine learning algorithms. It also provides feature extraction, transformation, dimensionality reduction, and selection as examples of featurization.
The previous studies of CKD prediction have not used big data platforms to solve this problem. The goal of this work is to predict CKD using hybrid ML techniques based on Apache Spark to predict CKD. Our contribution can be summarized as follows:
Developing hybrid ML techniques based on Apache Spark to predict CKD
Applying feature selection algorithms to select the important features from the dataset
Applying optimization techniques, including grid search with cross-validation to optimize ML algorithms to enhance performance
Applying different ML classification algorithms to both full features and the selected features
Applying ensemble learning such as Gradient-Boosted Trees based on Apache Spark to predict CKD.
The rest of this paper is structured as follows: Section 2 presents the previous studies to predict CKD. Section 3 presents the main stages of a developing system to predict CKD based on Apache Spark. Section 4 presents the experimental results. Finally, conclusions are presented in Section 5.
2. Related Works
Many authors have used different ML techniques for the diagnosis and prediction of chronic kidney disease as shown in Table 1.
Table 1.
Related works for prediction of CKD.
| REF | Year | Models | Feature selection methods | Dataset |
|
| ||||
| [22] | 2021 | SVM, KNN, DT, and RF | Recursive feature elimination (RFE) | CKD dataset |
| [20] | 2020 | ANN, C5.0, and LR | CFS, Lasso, and | CKD dataset |
| LSVM, KNN, and RF | Wrapper method | |||
| [23] | 2020 | RF, SVM, NB, and LR | RF-FS, FS, FES, BS, and BES | CKD dataset |
| [24] | 2020 | An ensemble of decision tree models | Cost-sensitive ensemble | CKD dataset |
| Feature ranking | ||||
| [25] | 2020 | Bagging and random subspace | No | CKD dataset |
| Methods based on KNN | ||||
| NB and DT | ||||
| [26] | 2020 | Decision Table, J48 | Genetic search algorithm | CKD dataset |
| MLP and NB | ||||
| [27] | 2019 | LR, RF, SVM, KNN | No | CKD dataset |
| NB and FNN | ||||
| A hybrid model LR and RF | ||||
| [28] | 2019 | Artificial neural network (ANN) and SVM | Correlation coefficients | CKD dataset |
| [29] | 2018 | NB and K-Star | No | CKD dataset |
| SVM | ||||
| J48 | ||||
| [30] | 2018 | AdaBoost and KNN | CFS | CKD dataset |
| NB and SVM | ||||
For example, in [27], the authors proposed a hybrid model that combines LR and RF to predict CKD disease. They compared their proposed model with six ML algorithms, LR, RF, SVM, KNN, Naive Bayes (NB), and feedforward neural network (FNN). Their proposed model has registered the highest accuracy at 99.83%. In [29], NB, K-Star, SVM, and J48 classifiers were used to predict CKD. Performance comparison was made using WEKA software. J48 algorithm had better performance with 99% accuracy than the other algorithms.
Some authors used ML algorithms with feature selection methods to predict CKD. In [22], the recursive feature elimination (RFE) feature selection method has been used to select the essential features from the chronic kidney disease (CKD) dataset. Four classification algorithms have been applied (SVM, KNN, DT, and RF) to both full features and selected features. The results showed that RF outperformed all other algorithms. In [20], the authors used chi-square, CFS, and Lasso feature selection to select the essential features from the database. They applied ANN, C5.0, LR, LSVM, KNN, and RF to both full features and the selected features. The results showed that LSVM with full features has registered the highest accuracy at 98.86%. In [23], five feature selection methods, Random Forest feature selection (RF-FS), forward selection (FS), forward exhaustive selection (FES), backward selection (BS), and backward exhaustive (BE), have been used to select the most important features from the database. Four ML algorithms, RF, SVM, NB, and LR, have been used to predict CKD. The results showed that RF with Random Forest feature selection had achieved the best performance with 98.8% accuracy. In [26], the genetic search algorithm has been used to select the most important features from the CKD dataset. Decision Table, J48, Multilayer Perceptron (MLP), and NB have been applied to both full features and the selected features. Using genetic search algorithm enhanced the performance. The MLP classifier has achieved the best performance and outperformed the other classifiers. In [30], the number of important features has been selected using a correlation-based feature selection (CFS). AdaBoost, KNN, NB, and SVM have been used to detect CKD. The proposed CFS with AdaBoost achieved the best performance at 98.1% accuracy. In [25], the authors used two ensembles techniques which are Bagging and Random Subspace methods and three base-learners, KNN, NB, and DT, to predict CKD. The random subspace has achieved the best performance than Bagging on KNN classifier.
Previous studies just applied ML techniques to study and analyze data about CKD; they did not use big data platforms. Therefore, this motivates us to use big data platform (Spark) to study and analyze data about CKD including hybrid approaches (feature selection methods with ML classification algorithms and feature selection methods with ensemble algorithms).
3. Methodology
The proposed system of predicting chronic kidney disease consists of two main approaches, as shown in Figure 1. The first approach uses feature selection methods to select the essential features from the chronic kidney disease datasets. The second approach applies ML techniques: DT, LR, RF, SVM, NB, and ensemble learning on the selected features and full features to predict CKD. The proposed system is composed of 6 steps: in the first step (data collection), the CKD dataset from the UCI machine learning repository will be used. In the second step (data preprocessing step), null values will be handled. In the third step, the feature methods will be used to select the essential features. In the fourth step, a grid search with stratified cross-validation is used to optimize the parameters of ML and ensemble learning techniques. Each step is described in detail in the following subsections.
Figure 1.
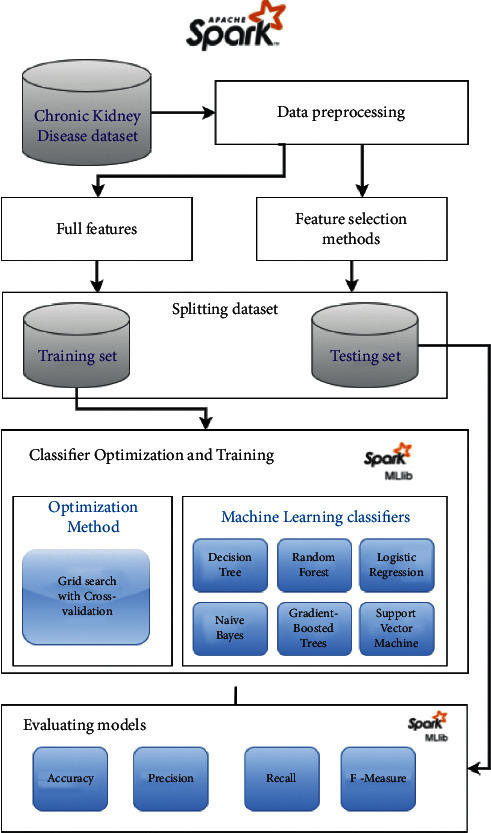
The steps of prediction CKD based on Apache Spark.
3.1. Data Collection
The chronic kidney disease (CKD) dataset used in this study was obtained from the UCI machine learning repository [31]. The CKD dataset includes 400 samples, 24 features, and 1 class label. The dataset contains 400 samples. The class label has two values: ckd (sample with CKD) and notckd (sample without CKD). The details of each feature are described in Table 2.
Table 2.
The CKD dataset description.
| Features | Explain |
|
| |
| age | Age |
| bp | Blood pressure |
| sg | Specific gravity |
| al | Albumin |
| su | Sugar |
| rbc | Red blood cells |
| pc | Pus cell |
| pcc | Pus cell clumps |
| ba | Bacteria |
| bgr | Blood glucose random |
| bu | Blood urea |
| sc | Serum creatinine |
| sod | Sodium |
| pot | Potassium |
| hemo | Hemoglobin |
| pcv | Packed cell volume |
| wc | White blood cell count |
| rc | Red blood cell count |
| htn | Hypertension |
| dm | Diabetes mellitus |
| cad | Coronary artery disease |
| appet | Appetite |
| pe | Pedal edema |
| ane | Anemia |
| class | Class |
3.2. Data Preprocessing
The dataset included outliers and noise. Therefore, it needs to be cleaned up and unblemished in a preprocessing stage. The preprocessing stage incorporated the estimation of the missing values and noise elimination, like outliers, normalization, and unbalanced data checking, because certain measures may be lost when patients are being tested, resulting in missing values. There were 158 completed cases in the dataset, with the remainder occurrences having missing values. Ignoring the record is the simplest way of dealing with the missing values, although this strategy is ineffective in small datasets. Instead of removing records, we can apply algorithms to estimate the missing data as an alternative approach. The missing values of nominal features have been filled by mode. The missing values of numerical features have been filled by mean.
3.3. Feature Selection Methods
The main benefits of using feature selection algorithms are determining the important features in the dataset. The classifier approach with feature selection produces better results and reduces the model's execution time. Relief-F and chi-squared feature selection method were used to select the subset of important features from the database. This study has applied two feature selection strategies based on Apache Spark.
3.4. Splitting the Dataset
The CKD datasets are split into 80% training set and 20% testing set. We used stratified cross-validation to train and optimize the models using the training set and the result of cross-validation is registered. We evaluated the models using the testing set, and the results of the testing set are registered.
3.5. Models' Optimization and Training
3.5.1. Optimization Methods
Grid search with stratified K-Fold cross-validation is used to optimize the models and tune the hyperparameters. The most common method for hyperparameter optimization is grid search. For each hyperparameter, the users must first define a set of values. The model then evaluates all possible values for each hyperparameter and chooses the one that provides the best performance.
K-Fold cross-validation: the dataset is divided into k folds of equal size. The training is done in k-1 groups, with the remaining time being used to test the classifiers. This procedure is repeated until each of the ten folds has been provided as a testing set. The performance of the classifiers is also measured for each k. Finally, depending on the average performance, the evaluation classifier is created.
3.5.2. Machine Learning Models
The classification models used in the research are as follows:
Decision tree (DT): it could be a supervised rule for learning in classification issues that contains a predefined target variable which is generally used. Decision tree works for each specific and continuous input and output variables. During this methodology, decision tree will be applied to each classification and regression issue that divides the population or sample into two or additional same sets known as subpopulation supporting the foremost necessary splitter within the input variable [34].
Random forest (RF): it is a type of supervised ML technique. Basically, it accumulates a lot of trees and integrates them for more accurate prediction [23].
Logistic regression (LR): it solved binary classification problems. A logistic or sigmoid function is used in LR to predict the probabilities of various labels for an unlabeled observation [35].
Support vector machine (SVM): it is a type of supervised ML technique. It segregates dataset into classes using the hyperplane [22].
Naïve Bayes (NB): the Bayes theorem is used to train a classifier in the Nave Bayes algorithm. In other words, it is a probabilistic classifier that has been trained using the Nave Bayes algorithm. It calculates a probability distribution over a set of classes for a given observation [29].
Gradient-Boosted Trees (GBTs): it is also possible to train an ensemble of decision trees using the Gradient-Boosted Trees (GBTs) algorithm. However, each decision tree is trained sequentially. This makes use of the previously trained tree information to optimize each new tree. As a result, the model improves with every new tree. Since GBT trains one tree at a time, it can take longer time to train a model using GBT. In addition, if many trees are used in an ensemble, it is prone to overfitting. In a GBT ensemble, each tree can, however, be shallow, making it easier to train. Gradient boosting is a technique for iteratively training a series of decision trees. On each iteration, the method predicts the label of each training sample using the current ensemble and then compares the prediction to the true label [36].
3.6. Evaluating the Models
As shown in Equations 1-4, the models are evaluated using four standard metrics: accuracy, precision, recall, and F1-score, where TP stands for true positive, TN stands for true negative, FP stands for false positive, and FN stands for false negative.
| (1) |
| (2) |
| (3) |
| (4) |
4. Experiments and Results
This section discusses the results of applying chi-square and Relief-F to the dataset to select the most important features. Also, it discusses the performance of cross-validation and the testing results of applying ML algorithms, SVM, LR, NB, RF, DT, and GBT Classifier, to the full features and the selected features. In addition, it demonstrates the best values of parameters for each ML algorithm that was optimized by grid search. Two feature selection methods were used; the CKD dataset was split into 80% training set and 20% testing set. The cross-validation results were registered for the training set, and the testing results were registered for the testing set. ML algorithms and features selection methods were implemented using PySpark.
4.1. Results of Chi-Square Feature Selection Method and ML Algorithms
In this subsection, the essential features were selected by chi-square algorithm to pass into ML models for predicting CKD. The 12 most important features which have the highest scores and were thus used to predict CKD chi-square are wc, bgr, bu, sc, pcv, al, haem, age, su, htn, dm, and bp, as shown in Figure 2. It can be noticed that wc has the highest score at 12733.72, while bp has the lowest score at 80.02. The second highest score is registered by bgr at 2428.327. Sc and pcv have the same score at 354.410 and 324.706, respectively. Also, htn and dm have approximately the same score at 86.29 and 80.44, respectively. Table 3 displays the scores of all features that chi-square has selected. The highest score is registered by wc at 12733.72, while the lowest is registered by sg at 0.0050.
Figure 2.
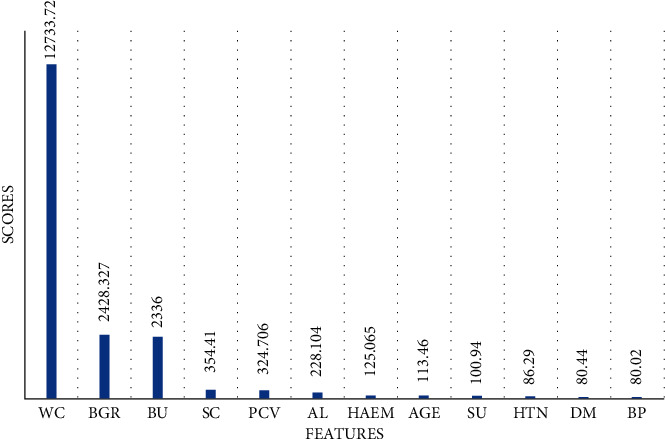
The important features selected by chi-square.
Table 3.
The scores of all features that are selected by chi-square.
| Features | Scores |
|
| |
| wc | 12 733.72 |
| bgr | 2428.327 |
| bu | 2336.00 |
| sc | 354.410 |
| pcv | 324.706 |
| al | 228.104 |
| haem | 125.065 |
| age | 113.460 |
| su | 100.94 |
| htn | 86.29 |
| dm | 80.44 |
| bp | 80.02 |
| pe | 45.10 |
| ane | 35.611 |
| sod | 28.793 |
| pcc | 24.075 |
| rc | 20.84 |
| cad | 19.93 |
| pc | 14.16 |
| ba | 12.58 |
| appe | 12.58 |
| rbc | 9.41 |
| pot | 4.07 |
| sg | 0.0050 |
The performance of cross-validation and the testing results of applying ML to the selected features by chi-square are described in Table 4. For cross-validation result, RF registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB has registered the lowest performance (AC = 81%, PR = 85%, RE = 82%, FS = 82%). LR and SVM have the same performance (AC = 97%, PR = 97%, RE = 97%, FS = 97%). For the testing results, SVM registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB registered the lowest performance (AC = 82%, PR = 88%, RE = 82%, FS = 82%). The second highest performance is registered by LR (AC = 97%, PR = 98%, RE = 97%, FS = 97%).
Table 4.
The performance of ML with the features selected by chi-square.
| Models | Cross-validation performance | Test performance | ||||||
| AC | PR | RE | FS | ACC | PR | RE | FS | |
|
| ||||||||
| DT | 97 | 98 | 98 | 98 | 92 | 93 | 93 | 93 |
| RF | 100 | 100 | 100 | 100 | 95 | 95 | 95 | 95 |
| LR | 97 | 97 | 97 | 97 | 97 | 98 | 97 | 97 |
| SVM | 97 | 97 | 97 | 97 | 100 | 100 | 100 | 100 |
| NB | 81 | 85 | 82 | 82 | 82 | 88 | 82 | 82 |
| GBT Classifier | 98 | 98 | 98 | 98 | 95 | 95 | 95 | 95 |
For optimization ML models, some of values of parameters are adapted and the best setting of ML's parameters is shown in Table 5.
Table 5.
The best values of ML's parameters are applied to the features selected by chi-square.
| Model | Parameters | Values |
|
| ||
| DT | Impurity | Gini |
| maxDepth | 3 | |
| maxBins | 10 | |
|
| ||
| RF | Impurity | Gini |
| maxDepth | 6 | |
| maxBins | 32 | |
|
| ||
| LR | regParam | 0.8 |
| maxIter | 20 | |
|
| ||
| SVM | regParam | 0.01 |
| maxIter | 100 | |
| NB | Smoothing | 0.2 |
|
| ||
| GBT Classifier | maxDepth | 2 |
| maxBins | 60 | |
4.2. Results of Relief-F Feature Selection Method and ML Algorithms
In this subsection, the essential features were selected by Relief-F algorithm to pass into Ml models for predicting CKD. The 12 most important features which have the highest weights selected by Relief-F and were used to predict CKD are shown in Figure 3. It can be noticed that rbc has the highest weight at 0.4551, while appe has the lowest weight at 0.062875. The second highest weight is registered by haem at 0.365745. Al and dm have approximately the same weights at 0.257775 and 0.24085, respectively.
Figure 3.
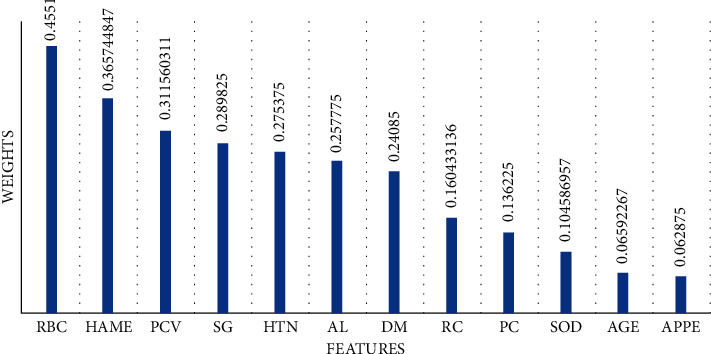
The weights of the most essential selected by Relief-F.
Table 6 displays weights of all features that are selected by Relief-F. The highest weight is registered by rbc at 0.4551, while the lowest weight is registered by bp at -0.01584. The performance of cross-validation and the testing results of applying ML to the features selected by Relief-F are described in Table 7. For cross-validation results, DT, RF, and GBT Classifier registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB registered the lowest performance (AC = 88%, PR = 89%, RE = 89%, FS = 89%). LR and SVM have the same performance (AC = 99%, PR = 99%, RE = 99%, FS = 99%).
Table 6.
The performance of ML with the features selected by Relief-F.
| Models | Cross-validation performance | Test performance | ||||||
| AC | PR | RE | FS | AC | PR | RE | FS | |
|
| ||||||||
| DT | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| RF | 100 | 100 | 100 | 100 | 98 | 99 | 99 | 99 |
| LR | 99 | 99 | 99 | 99 | 98 | 99 | 99 | 99 |
| SVM | 99 | 99 | 99 | 99 | 98 | 99 | 99 | 99 |
| NB | 88 | 89 | 89 | 89 | 95 | 95 | 95 | 95 |
| GBT Classifier | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
Table 7.
The best values of ML's parameters which are applied to the features selected by Relief-F.
| Model | Parameters | Values |
|
| ||
| DT | Impurity | Gini |
| maxDepth | 4 | |
| maxBins | 32 | |
|
| ||
| RF | Impurity | Gini |
| maxDepth | 5 | |
| maxBins | 32 | |
|
| ||
| LR | regParam | 0.1 |
| maxIter | 20 | |
|
| ||
| SVM | regParam | 0.01 |
| maxIter | 100 | |
| NB | Smoothing | 0.1 |
|
| ||
| GBT Classifier | maxDepth | 4 |
| maxBins | 20 | |
For the testing results, DT and GBT Classifier registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB registered the lowest performance (AC = 95%, PR = 95%, RE = 95%, FS = 95%). LR and SVM have the same performance (AC = 98%, PR = 99%, RE = 99%, FS = 99%).
For optimization ML models, some of values of parameters are adapted and the best setting of ML's parameters is shown in Table 8.
Table 8.
The weights of all features that are selected by Relief-F.
| Features | Weights |
|
| |
| rbc | 0.455 1 |
| haem | 0.365 745 |
| pcv | 0.311 56 |
| sg | 0.289 825 |
| htn | 0.275 375 |
| al | 0.257 775 |
| dm | 0.240 85 |
| rc | 0.160 433 |
| pc | 0.136 225 |
| sod | 0.104 587 |
| Age | 0.065 923 |
| appe | 0.062 875 |
| pe | 0.056 825 |
| su | 0.031 65 |
| bgr | 0.029 549 |
| ane | 0.027 |
| bu | 0.022 733 |
| sc | 0.015 806 |
| pcc | 0.015 675 |
| wc | 0.006 426 |
| ba | −0.000 12 |
| pot | −0.004 11 |
| cad | −0.011 97 |
| bp | −0.015 84 |
4.3. The Performance of ML with Full Features
Table 9 presents the result of cross-validation and the testing of applying ML to full features. Overall, RF achieved the best performance for cross-validation and the testing results. For cross-validation results, RF registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB has the lowest performance (AC = 84%, PR = 88%, RE = 84%, FS = 84%). LR, SVM, and GBT Classifier have the same performance (AC = 99%, PR = 99%, RE = 99%, FS = 99%). For the testing results, RF and SVM registered the highest performance (AC = 100%, PR = 100%, RE = 100%, FS = 100%), while NB has the lowest performance (AC = 87%, PR = 91%, RE = 88%, FS = 88%). For optimization ML models, some of values of parameters are adapted and the best setting of ML's parameters is shown in Table 10.
Table 9.
The performance of ML with full features.
| Models | Cross-validation performance | Test performance | ||||||
|---|---|---|---|---|---|---|---|---|
| AC | PR | RE | FS | AC | PR | RE | FS | |
| DT | 98.43 | 98 | 98 | 98 | 95 | 95 | 95 | 95 |
| RF | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| LR | 99 | 99 | 99 | 99 | 98 | 99 | 99 | 99 |
| SVM | 99 | 99 | 99 | 99 | 100 | 100 | 100 | 100 |
| NB | 84 | 88 | 84 | 84 | 87 | 91 | 88 | 88 |
| GBT Classifier | 99 | 99 | 99 | 99 | 95 | 95 | 95 | 95 |
Table 10.
The best values of ML's parameters which are applied to full features.
| Model | Parameters | Values |
|
| ||
| DT | Impurity | Gini |
| maxDepth | 4 | |
| maxBins | 10 | |
|
| ||
| RF | Impurity | Gini |
| maxDepth | 7 | |
| maxBins | 32 | |
|
| ||
| LR | regParam | 0.3 |
| maxIter | 10 | |
|
| ||
| SVM | regParam | 0.01 |
| maxIter | 1000 | |
| NB | Smoothing | 0.2 |
|
| ||
| GBT Classifier | maxDepth | 2 |
| maxBins | 60 | |
4.4. Discussion
Table 11 presents models that have achieved the highest cross-validation results. The performance of cross-validation of applying ML to the features selected by Relief-F has achieved the best value by three models: DT, RF, and GBT Classifiers. In comparisons, the cross-validation performance of applying ML to full features and features selected by chi-square has achieved the best value by 1 model: RF.
Table 11.
Best models for cross-validation results.
| Best models | Features | Measure methods | |||
| AC | PR | RE | FS | ||
|
| |||||
| RF | Full features | 100 | 100 | 100 | 100 |
| RF | Features selected by chi-square | 100 | 100 | 100 | 100 |
| DT | Features selected by Relief-F | 100 | 100 | 100 | 100 |
| RF | Features selected by Relief-F | 100 | 100 | 100 | 100 |
| GBT Classifier | Features selected by Relief-F | 100 | 100 | 100 | 100 |
Table 12 presents the best models for the testing results. The performance of testing applying ML to the features selected by Relief-F has achieved the best value by two models: DT and GBT Classifiers. The testing performance of applying ML to full features has achieved the best value by two models: RF and SVM Classifiers. However, the testing performance of applying ML to the features selected by chi-square has achieved the best value by 1 model: SVM Classifier.
Table 12.
Best models for the testing results.
| Best models | Features | Measure methods | |||
| AC | PR | RE | FS | ||
|
| |||||
| SVM | Full features | 100 | 100 | 100 | 100 |
| RF | Full features | 100 | 100 | 100 | 100 |
| SVM | Features selected by chi-square | 100 | 100 | 100 | 100 |
| DT | Features selected by Relief-F | 100 | 100 | 100 | 100 |
| GBT Classifier | Features selected by Relief-F | 100 | 100 | 100 | 100 |
The results showed that SVM, DT, and GBT Classifier with the selected features have achieved the best performance. Overall, the performance with Relief-F feature selection is better than chi-square feature selection and full features.
Table 13 presents the comparison of performance between the previous studies and our work on the same dataset. In our work, the Relief-F feature selection methods have achieved the best performance for the testing results and cross-validation results using DT and GBT Classifier compared to the other existing works [23, 24, 26, 27, 30]. Also, our work is different from the other existing works [22, 25] because it registered the results for both the training set and the testing set, and it has achieved the best performance.
Table 13.
The comparison of performance between the previous studies and our work on the same dataset.
| REF | Feature selection methods | The best model | Dataset | Result |
|
| ||||
| [22] | RFE | RF | CKD dataset | AC = 100% |
| PR = 100% | ||||
| RE = 100% | ||||
| FS = 100% | ||||
| [27] | No | A hybrid model LR and RF | CKD dataset | AC = 99.94% |
| E = 99.84% | ||||
| S = 99.80% | ||||
| [30] | CFS | AdaBoost based on KNN | CKD dataset | AC = 98.1% |
| PR = 98% | ||||
| RE = 98% | ||||
| FS = 98% | ||||
| [23] | Rffs, FS, FES, BS, BES | RF | CKD dataset | AC = 98.825% |
| RE = 98.04% | ||||
| [24] | Cost-sensitive ensemble feature ranking | An ensemble of decision tree models | CKD dataset | AC = 97.27% |
| PRC = 99.44% | ||||
| RE = 96.25% | ||||
| FS = 97.68% | ||||
| [25] | No | Random subspace-based KNN | CKD dataset | AC = 100% |
| RE = 100% | ||||
| [26] | Genetic search algorithm | Multilayer perceptron | CKD dataset | AC = 99.75% |
| Our work | Relief-F | DT | CKD dataset | Cross-validation result AC = 100%, PRC = 100%, RRE = 100% FS = 100% result of testing AC = 100%, PRC = 100%, RRE = 100%, FS = 100% |
| GBT Classifier | CKD dataset | Cross-validation result AC = 100%, PRC = 100%, RRE = 100%, FS = 100%; result of testing AC = 100%, PRC = 100%, RRE = 100%, FS = 100% | ||
5. Conclusion
In this paper, the hybrid ML techniques integrating feature selection methods and classification ML algorithms based on big data platforms (Apache Spark) were used to predict CKD. Relief-F and chi-squared feature selection techniques were used to select the important features from the dataset. ML algorithms, DT, LR, NB, RF, SVM, and GBT Classifier as ensemble learning algorithm, were applied to benchmark chronic kidney disease dataset. Also, they were applied to the full features and to the selected features. Grid search with cross-validation was used to optimize the parameters of ML. In addition. Four methods of evaluation, accuracy, precision, recall, and F1-measure, were applied to validate the results and the results of cross-validation and the testing data were registered. The results showed that SVM, DT, and GBT Classifier with the selected features have achieved the best performance. Overall, the performance of Relief-F feature selection is better than that achieved by chi-square feature selection and the full features.
Acknowledgments
The authors would want to express their gratitude to the UCI machine learning repository for providing CKD dataset.
Data Availability
Chronic kidney disease dataset is downloaded from https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease.
Conflicts of Interest
All authors declare that they have no conflicts of interest.
References
- 1.Manogaran G., Lopez D. Health data analytics using scalable logistic regression with stochastic gradient descent. International Journal of Advanced Intelligence Paradigms . 2018;10(1-2):118–132. doi: 10.1504/ijaip.2018.089494. [DOI] [Google Scholar]
- 2.Hu H., Wen Y., Chua T. S., Li X. Toward scalable systems for big data analytics: a technology tutorial. IEEE access . 2014;2:652–687. doi: 10.1109/access.2014.2332453. [DOI] [Google Scholar]
- 3.Apache Hadoop. 2021. https://hadoop.apache.org/
- 4.Kafka A. Apache Kafka. 2021. https://kafka.apache.org/
- 5.Storm A. Apache Storm. 2021. https://storm.apache.org/
- 6.Beam A. L., Kohane I. S. Big data and machine learning in health care. JAMA . 2018;319(13):1317–1318. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 7.Nair L. R., Shetty S. D., Shetty S. D. Applying spark based machine learning model on streaming big data for health status prediction. Computers & Electrical Engineering . 2018;65:393–399. doi: 10.1016/j.compeleceng.2017.03.009. [DOI] [Google Scholar]
- 8.Ali A. A. Stroke prediction using distributed machine learning based on Apache spark. Stroke . 2019;28(15):89–97. [Google Scholar]
- 9.World Health Organization. Preventing Chronic Diseases: A Vital Investment . Geneva, Switzerland: World Health Organization; 2005. [Google Scholar]
- 10.Bikbov B., Perico N., Remuzzi G. Disparities in chronic kidney disease prevalence among males and females in 195 countries: analysis of the global burden of disease 2016 study. Nephron . 2018;139(4):313–318. doi: 10.1159/000489897. [DOI] [PubMed] [Google Scholar]
- 11.Disease K. Improving global outcomes (kdigo) transplant work group. kdigo clinical practice guideline for the care of kidney transplant recipients. American Journal of Transplantation . 2009;9(3):S1–S155. doi: 10.1111/j.1600-6143.2009.02834.x. [DOI] [PubMed] [Google Scholar]
- 12.Cdc. Chronic Kidney Disease in the united states. 2021. https://www.cdc.gov/kidneydisease/publications-resources/ckd-national-facts.html .
- 13.Deng L., Li X. Machine learning paradigms for speech recognition: an overview. IEEE Transactions on Audio Speech and Language Processing . 2013;21(5):1060–1089. doi: 10.1109/tasl.2013.2244083. [DOI] [Google Scholar]
- 14.Huang M. Q., Ninić J., Zhang Q. B. Bim, machine learning and computer vision techniques in underground construction: current status and future perspectives. Tunnelling and Underground Space Technology . 2021;108 doi: 10.1016/j.tust.2020.103677.103677 [DOI] [Google Scholar]
- 15.Oza P., Sharma P., Patel S. Proceedings of the Second International Conference on Computing, Communications, and Cyber-Security . New York, NY, USA: Springer; 2021. Machine learning applications for computer-aided medical diagnostics. [DOI] [Google Scholar]
- 16.Bikmukhametov T., Jäschke J. Combining machine learning and process engineering physics towards enhanced accuracy and explainability of data-driven models. Computers & Chemical Engineering . 2020;138 doi: 10.1016/j.compchemeng.2020.106834.106834 [DOI] [Google Scholar]
- 17.Palma-Mendoza R. J., Rodriguez D., Marcos L. D. Distributed relieff-based feature selection in spark. Knowledge and Information Systems . 2018;57(1):1–20. doi: 10.1007/s10115-017-1145-y. [DOI] [Google Scholar]
- 18.Nassar M., Safa H., Mutawa A. A., Helal A., Gaba I. Chi squared feature selection over Apache spark. Proceedings of the 23rd International Database Applications & Engineering Symposium; June 2019; Athens Greece. pp. 1–5. [DOI] [Google Scholar]
- 19.Charleonnan A., Fufaung T., Niyomwong T., Chokchueypattanakit W., Suwannawach S., Ninchawee N. Predictive analytics for chronic kidney disease using machine learning techniques. Proceedings of the 2016 management and innovation technology international conference (MITicon); October 2016; Bang-San, Thailand. [DOI] [Google Scholar]
- 20.Chittora P., Chaurasia S., Chakrabarti P., et al. Prediction of chronic kidney disease-a machine learning perspective. IEEE Access . 2021;9(17312):p. 17334. doi: 10.1109/access.2021.3053763. [DOI] [Google Scholar]
- 21.Spark A. Apache Spark . 2021. https://spark.apache.org/ [Google Scholar]
- 22.Senan E. M., Adhaileh M. H. A., Alsaade F. W., et al. Diagnosis of chronic kidney disease using effective classification algorithms and recursive feature elimination techniques. Journal of Healthcare Engineering . 2021;2021:10. doi: 10.1155/2021/1004767.1004767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdullah A. A., Hafidz S. A., Khairunizam W. Performance comparison of machine learning algorithms for classification of chronic kidney disease (ckd) Journal of Physics: Conference Series . 2020;1529(5) doi: 10.1088/1742-6596/1529/5/052077.052077 [DOI] [Google Scholar]
- 24.Ali S. I., Ali B., Hussain J., et al. Cost-sensitive ensemble feature ranking and automatic threshold selection for chronic kidney disease diagnosis. Applied Sciences . 2020;10(16):p. 5663. doi: 10.3390/app10165663. [DOI] [Google Scholar]
- 25.Jongbo O. A., Adetunmbi A. O., Ogunrinde R. B., Ajisafe B. B. Development of an ensemble approach to chronic kidney disease diagnosis. Scientific African . 2020;8 doi: 10.1016/j.sciaf.2020.e00456.e00456 [DOI] [Google Scholar]
- 26.Jena L., Patra B., Nayak S., Mishra S., Tripathy S. Intelligent and Cloud Computing . New York, NY, USA: Springer; 2021. Risk prediction of kidney disease using machine learning strategies; pp. 485–494. [DOI] [Google Scholar]
- 27.Qin J., Chen L., Liu Y., Liu C., Feng C., Chen B. A machine learning methodology for diagnosing chronic kidney disease. IEEE Access . 2019;8 doi: 10.1109/ACCESS.2019.2963053.20991 [DOI] [Google Scholar]
- 28.Almansour N. A., Syed H. F., Khayat N. R., et al. Neural network and support vector machine for the prediction of chronic kidney disease: a comparative study. Computers in Biology and Medicine . 2019;109:101–111. doi: 10.1016/j.compbiomed.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Avci E., Karakus S., Ozmen O., Avci D. Performance comparison of some classifiers on chronic kidney disease data. Proceedings of the 2018 6th International Symposium on Digital Forensic and Security (ISDFS); March 2018; Antalya, Turkey. pp. 1–4. [DOI] [Google Scholar]
- 30.Wibawa M. S., Maysanjaya I. M. D., Putra I. M. A. W. Boosted classifier and features selection for enhancing chronic kidney disease diagnose. Proceedings of the 2017 5th international conference on cyber and IT service management (CITSM); August 2017; Denpasar, Indonesia. pp. 1–6. [DOI] [Google Scholar]
- 31.Repository U. M. L. Chronic kidney disease data set. 2021. https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease .
- 32.Kononenko I. Estimating attributes: analysis and extensions of relief. Proceedings of the European conference on machine learning; April 1994; Catania, Italy. pp. 171–182. [DOI] [Google Scholar]
- 33.Canedo V. B., Remeseiro B. Sanchez-maro no, n., and alonso-betanzos. Transactions on Computational Collective Intelligence XX . 2017;9420:78–98. [Google Scholar]
- 34.Chaurasia V., Pal S., Tiwari B. Chronic kidney disease: a predictive model using decision tree. International Journal of Engineering Research and Technology . 2018;11(11):1781–1794. [Google Scholar]
- 35.Haq A. U., Li J. P., Memon M. H., Nazir S., Sun R. A hybrid intelligent system framework for the prediction of heart disease using machine learning algorithms. Mobile Information Systems . 2018;2018:21. doi: 10.1155/2018/3860146.3860146 [DOI] [Google Scholar]
- 36.Shi H., Wang H., Huang Y., Zhao L., Qin C., Liu C. A hierarchical method based on weighted extreme gradient boosting in ecg heartbeat classification. Computer Methods and Programs in Biomedicine . 2019;171:1–10. doi: 10.1016/j.cmpb.2019.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Chronic kidney disease dataset is downloaded from https://archive.ics.uci.edu/ml/datasets/chronic_kidney_disease.


