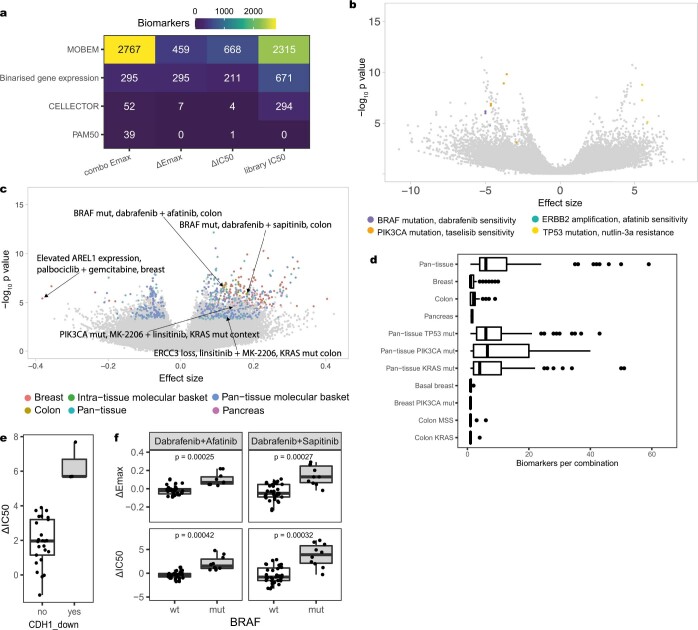
Extended Data Fig. 4. Landscape of biomarkers I.
a, Heatmap showing the distribution of 8,078 significant biomarkers across four inputs and four feature types. MOBEM: binary matrix of mutations, copy number alterations and methylations present in cell lines (see Methods for feature selection). b, Volcano plot of single-agent response biomarker associations tested (library IC50; n = 1,922,552; significant and large-effect biomarkers n = 3,280), with select statistically significant large-effect size biomarkers showing known single agent examples highlighted, namely BRAF mutation and dabrafenib sensitivity (purple), ERBB2 amplification and afatinib sensitivity (turquoise), PIK3CA mutation and taselisib sensitivity (orange), and TP53 mutation and resistance to nutlin-3a (yellow). Biomarkers identified using ANOVA test, p ≤ 0.001, FDR ≤ 5%, Glass deltas for positive and negative populations both ≥ 1. c, Volcano plot of biomarkers tested for associations with ΔEmax (n = 2,006,328), with significant and large-effect biomarkers (n = 761) coloured by analysis type. Examples discussed in the text and selected outliers are labelled. Biomarkers identified using ANOVA test, p ≤ 0.001, FDR ≤ 5%, Glass deltas for positive and negative populations both ≥ 1. d, Number of significant and large-effect biomarkers found per combination per context. Median and interquartile range (IQR). e, ΔIC50 drug combination response for irinotecan+AZD7762 in pancreatic cell lines with low expression of CDH1. n = 3 CDH1_down, n = 27 not CDH1_down. ANOVA, p ≤ 0.001, FDR ≤ 5%. Median and IQR. f, Drug combination responses for dabrafenib paired with EGFR inhibitors afatinib or sapitinib in BRAF wild-type (wt; n = 36) and mutant (mut, n = 11) colon cell lines. ΔEmax and ΔIC50 were averaged across replicates and highest response between anchor concentrations is reported. Two-sided Welch’s t-test. Median and IQR.