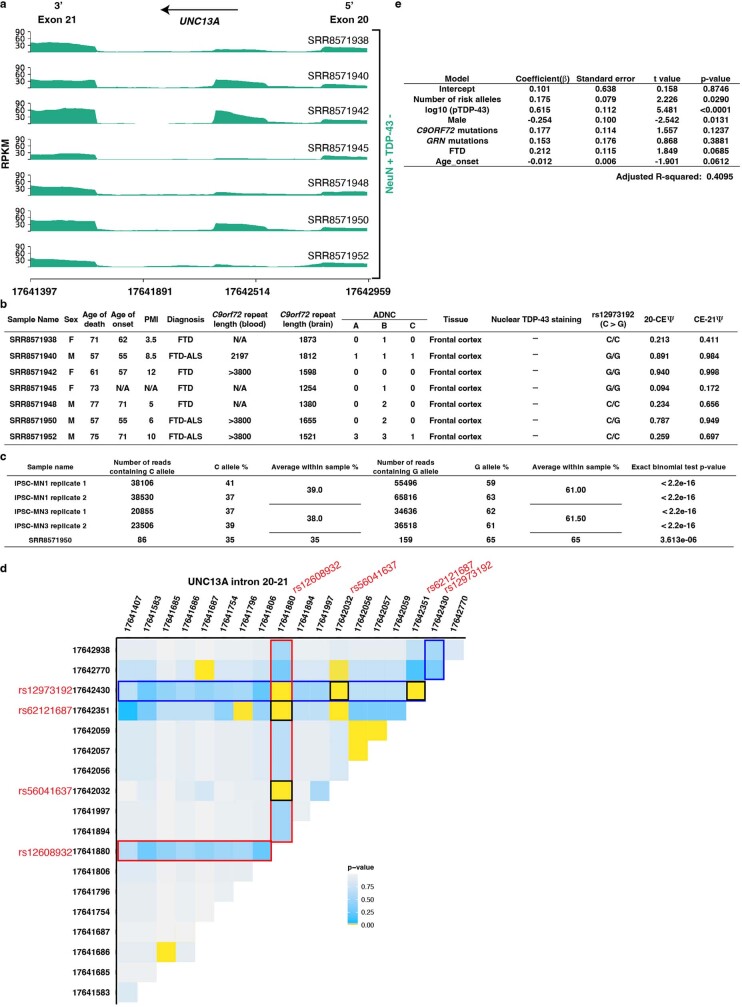
Extended Data Fig. 7. Levels of UNC13A cryptic exon inclusion are influenced by the number of risk haplotypes.
(a) Visualization of RNA-Seq alignment between exon 20 and exon 21 of UNC13A. RNA-Seq libraries were generated from TDP-43-negative neuronal nuclei as described in Fig. 1a. (b) Samples that are heterozygous (C/G) or homozygous (G/G) at rs12973192 have higher relative inclusion (Ψ) of the cryptic exon except for SRR8571945. Information about the patients were obtained from Liu et al. 12. (c) Percentages of C and G alleles in the UNC13A spliced variants in TDP-43 depleted iPSC-MNs and SRR8571950 neuronal nuclei. Exact binomial test was done for each replicate to test whether the observed difference in percentages differ from what was expected if both alleles are equally included in the cryptic exon. (d) rs56041637 and rs62121687 are in strong linkage disequilibrium with both GWAS hits in intron 20-21 of UNC13A (Method). Along the axes of the heatplot are all loci that show variation among the 297 patients from Answer ALS in July 2020. Each tile represents the p-value from the corresponding Chi-Square test. P-value < 0.05 are shown in yellow and others are shown in blue or gray. Red and blue blocks highlight the associations of rs12608932 and rs12973192 with other genetic variants in intron 20-21 respectively. Significant associations common to both are circled in black. (e) The summary results of multiple linear regression modeling the effects of the number of UNC13A risk alleles on the abundance of UNC13A cryptic exon inclusion measured by RT-qPCR. A multivariable model was derived adjusting for phosphorylated TDP-43 levels (pTDP-43), sex, known genetic mutations, disease types, and the age of onset. As shown in Extended Data Fig. 5c, pTDP-43 levels have a strong effect on the abundance of UNC13A cryptic exon inclusion. Normality of residuals is tested by Shapiro-Wilk normality test (p-value = 0.2014).