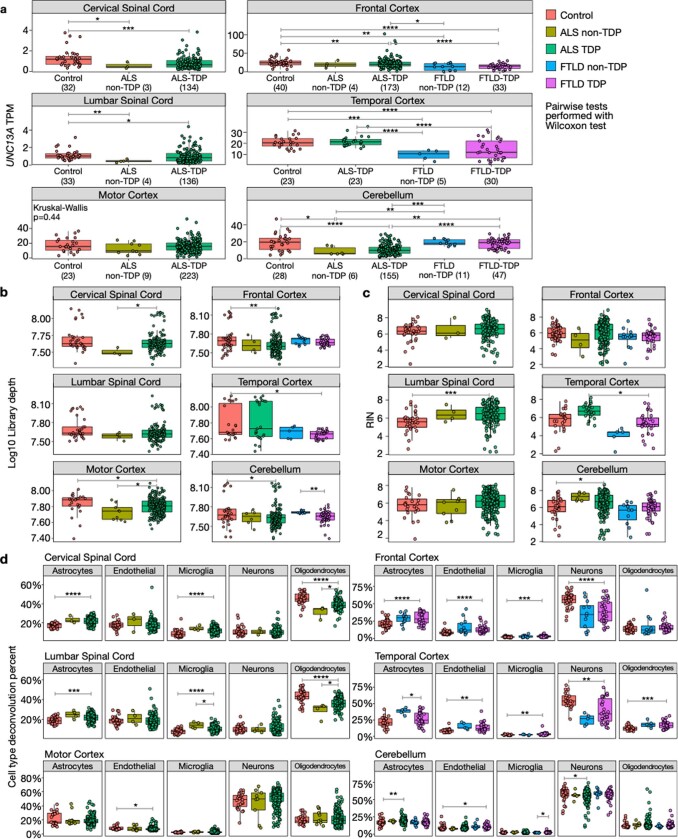
Extended Data Fig. 4. Sample technical factors in NYGC tissue samples do not vary in a systematic way.
(a) UNC13A expression across tissues and disease subtypes in the NYGC ALS Consortium RNA-seq dataset. Expression normalised as transcripts per million (TPM). Cortical regions have noticeably higher UNC13A expression than the spinal cord. (b) total RNA-seq library size (log10 scaled) (c) RNA integrity score (RIN) (d) Cell type decomposition across NYGC ALS Consortium RNA-seq dataset. While there are differences between tissues and disease-subtypes on these technical factors, specificity of UNC13A CE detection to tissues presumed to contain TDP-43 proteinopathy cannot be explained by these technical factors. Box plots (a–d): boundaries 25–75th percentiles; midline, median; whiskers, Tukey style. Wilcoxon test, significance levels reported as * (p < 0.05) ** (p < 0.01) *** (p < 0.001) **** (p < 0.0001).