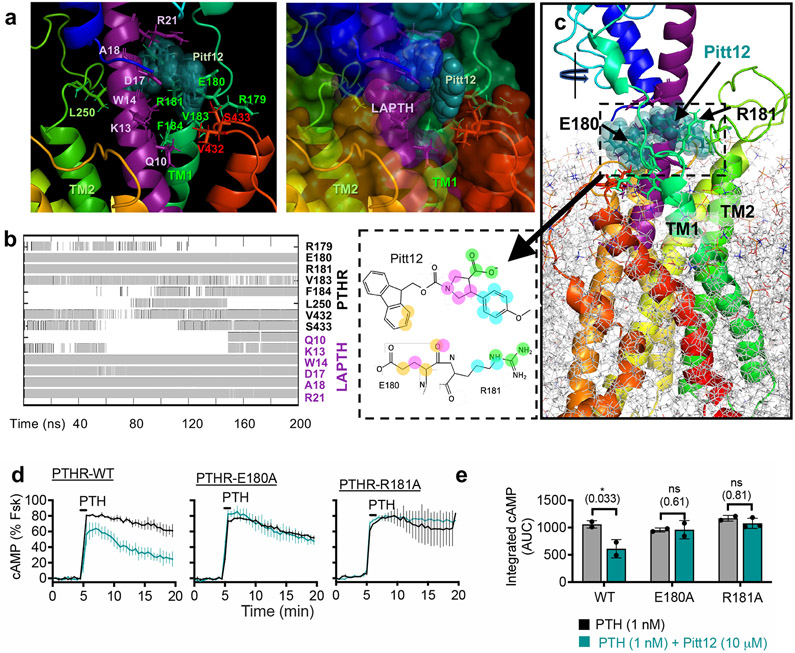
Figure 4. Refinement and validation of Pitt12 binding site in PTHR.
(a–c) Results from MD simulations (200 ns) of Pitt12 binding to active state, LA-PTH–bound PTHR (PDB 6NBF)12 embedded in membrane, showing predictive interactions (a), and time-evolution of Pitt12 engagement with the peptide ligand and PTHR residues (b), as well as the overall view of the simulation system and bound Pitt12 (c). Interaction is defined as existence of any heavy atom contact pairs between PTHR (or LA-PTH) and Pitt12 within distance < 4.0 Å. Inset of panel c shows 2D representations of closely interacting units, where same color circles (orange, magenta, cyan, or green) indicate interacting pairs of atoms between Pitt12 and E180/R181. As the initial pose of Pitt12- and LA-PTH-bound PTHR, we used the cryo-EM structure of LA-PTH-bound PTHR (PDB id: 6NBF), into which we inserted Pitt12 upon structural alignment with our snapshot from druggability simulations (which was adopted in the construction of the pharmacophore model as well as in virtual screening).
(d, e) Experimental validation of Pitt12 binding site in PTHR. PTH (1 nM)-induced cAMP generation time-courses (d) and corresponding integrated cAMP responses (e) from wild-type and receptor mutants carrying individual E180A or R181A mutation, in presence or absence of 10 μM Pitt12. Mean ± s.e.m. of N = 2 or 3 experiments with n = 39 (PTH) and n = 45 (PTH+Pitt12) cells examined for PTHR-WT, n = 46 (PTH) and n = 50 (PTH+Pitt12) cells examined for PTHR-E180A, and n = 40 (PTH) and n = 57 (PTH+Pitt12) cells examined for PTHR-R181A. P values were assessed by two-tailed Student’s t-test.