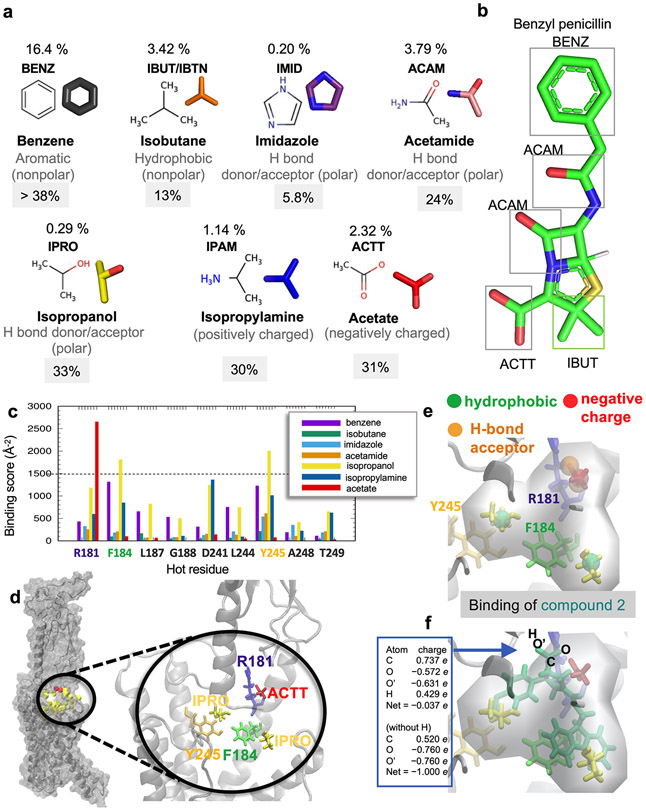
Extended Data Fig. 2. Probe molecules used in druggability simulations.
(a) Seven probe molecules used in PTHR druggability simulations. The structure of each probe is shown along with its 4-letter acronym, its name and main chemical properties. Also shown is the percentage of drugs containing those fragments based on the SMILES data of 2,453 approved drugs in DrugBank. (b) An example of a common drug, benzyl-penicillin, containing four types of fragments represented by the probe molecules. (c) Binding score profile for PTHR residues near Site 1 (labeled along the abscissa), evaluated for different probe molecules (bars in different colors). The binding score is defined as Σ (1/dki)2 where k is frame/snapshot index and dki is corresponding distance between the probe and the residue i summed over all snapshots n (12,000 compiled from six independent runs), provided that they make atom-atom contacts of dki < 4.0 Å. Residues with score above 1500/Å2 (dashed line) are selected as high affinity residues for each probe type: R1811.33b for acetate, and F1841.36b and Y2452.72b for isopropanol. (d) Close-up view of site 1 preferentially sampled by three probes (two isopropanol and an acetic acid, shown in yellow and red sticks respectively), and associated residues R1811.33b, F1841.36b and Y2452.72b (right). (e, f) Construction of pharmacophore model (PM) composed of a hydrogen bond acceptor, a negatively charged region, and two hydrophobic sites (spheres), based on the preferential positions observed in (c), and overlay of a hit compound Pitt12 (aquamarine sticks, extracted from the ZINC database) and the PM. The box on the left in panel (f) shows the partial charges for the carboxylate group atoms O, C, O’, and H in Pitt12 in its protonated and deprotonated forms. In line with the use of acetate probe molecule to construct the PM, we used the deprotonated form in further analyses and simulations.