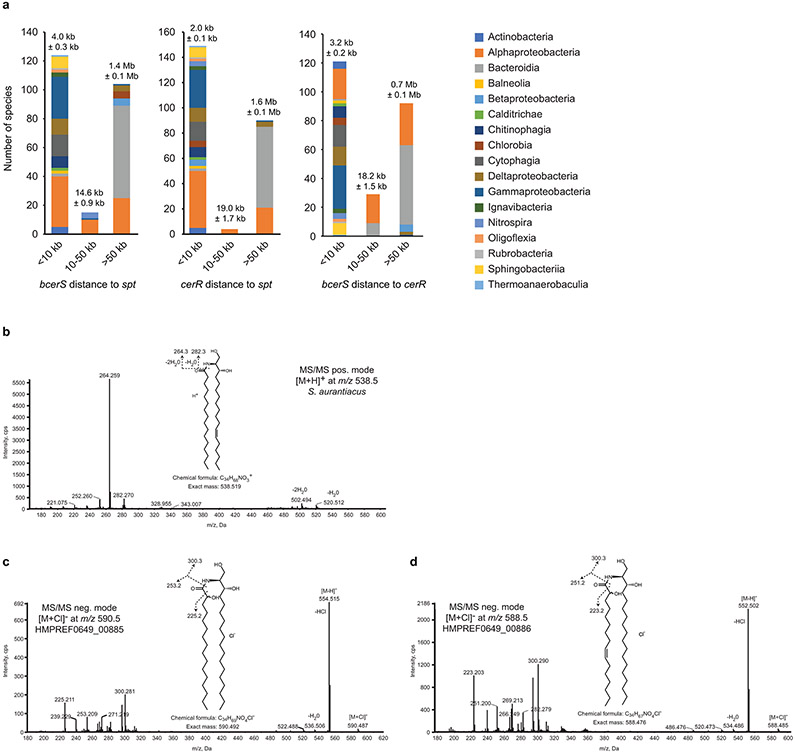
Extended Data Fig. 7. Ceramide synthesis orthologues.
(a) Using the genomic coordinates acquired during the phylogenetic analysis of bacterial ceramide genes, the distances between spt, bcerS, and cerR were calculated. The histogram columns are colored by taxonomic class. The numbers above the columns are the average distance ± the standard error of the mean. (b) MS/MS analysis of ceramides from S. aurantiacus confirm that the desaturation occurs on the long-chain base. (c) MS/MS analysis of ceramides produced by bCerS homologues from P. buccae (see Fig. 3f) show that HMPREF0649_00885 preferred fully saturated substrates, while HMPREF0649_00886 used a desaturated palmitoyl-CoA substrate.