Figure 4. A time course shows dramatic changes in stress responses with CNQX treatment.
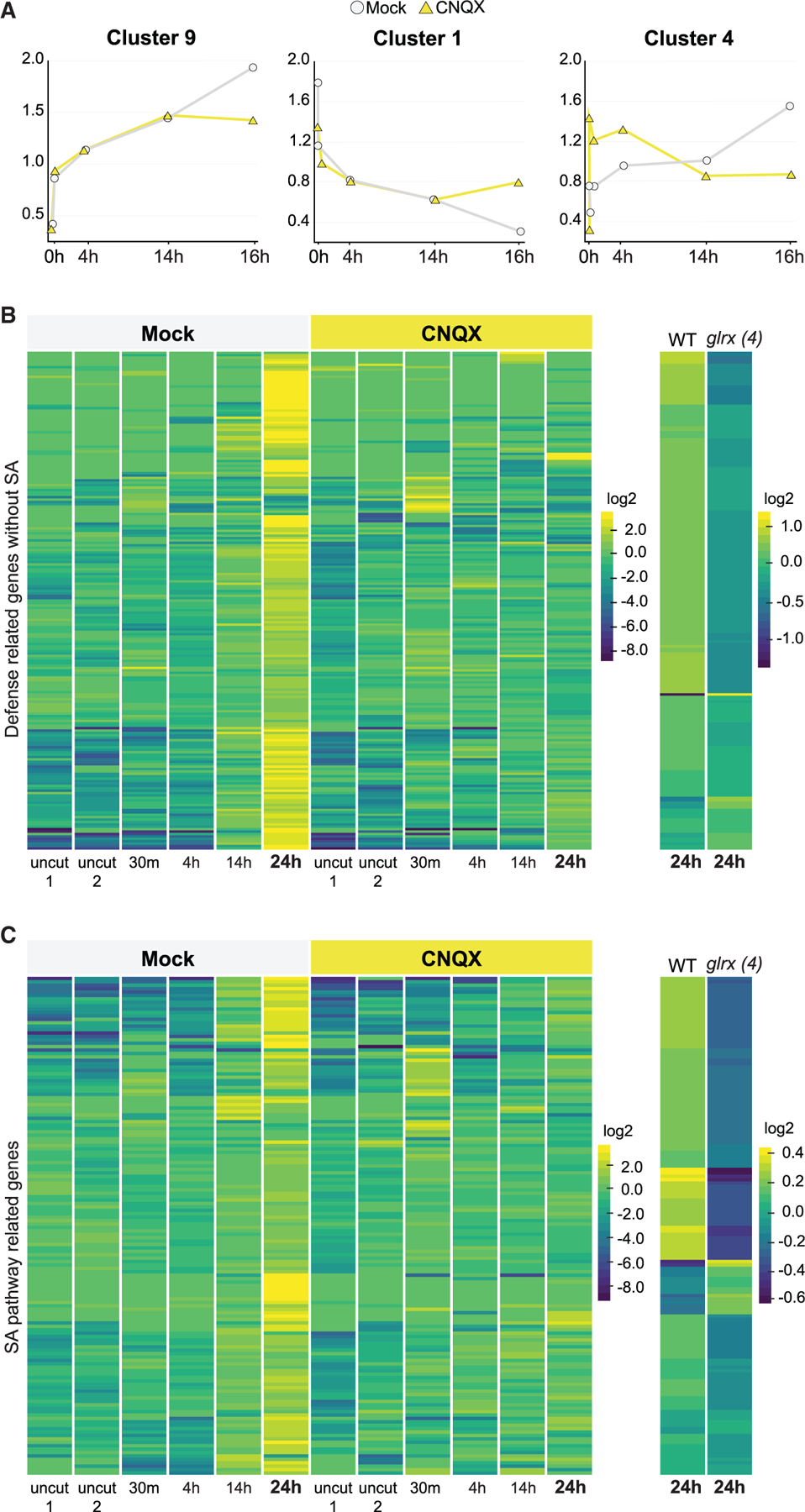
(A) Plots summarizing 3 of the 11 different patterns found among approximately 1,400 differentially regulated transcripts along a regeneration time course (0 min, 30 min, 4 h, 14 h, and 24 h), using ImpulseDE (see experimental model and subject details).
(B and C) Heatmaps of defense-annotated genes (B) and SA responsive genes (C) that are upregulated in regeneration at 24 h in untreated roots, showing a failure to upregulate the same genes in CNQX treatments and in the glrx4 mutant (glr1.2/1.4/2.2/3.3). For treatments, plants were grown on standard plates and then transferred to mock or CNQX plates (50 µM). Read counts were row normalized and log2 transformed. Col-0 versus glr1.2/1.4/2.2/3.3 mutant plants were profiled at 24 h after injury only and normalized separately. Genes in heatmaps are listed in Table S3.
