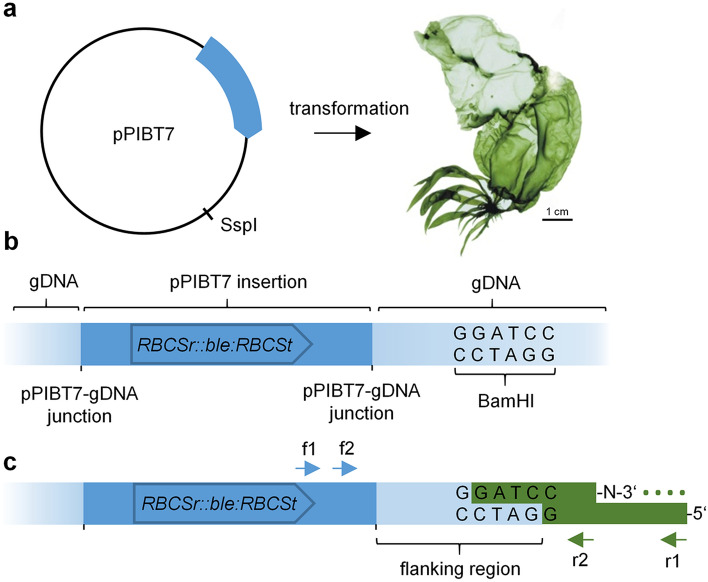
Fig. 1.
Schematic overview of adapter-ligation PCR approach. a For insertional mutagenesis, pPIBT7 carrying the phleomycin-resistance marker RBCSr::ble:RBCSt was linearized using SspI and transformed into Ulva, thereby creating genomic insertion sites as schematically shown in b. c Digestion of genomic DNA (gDNA) using BamHI (and BclI, not shown) leaves 5'GATC overhangs to which a compatible asymmetric adapter is ligated. The adapter 3′ amino-terminal group prevents DNA polymerase extension of the adapter sequence; consequently, the annealing site (dotted line) for the reverse primer (r1, arrow) only becomes available after the successful extension of the pPIBT7 insert specific primer f1 (arrow). A nested PCR using primers f2 and r2 further ensures the specific amplification of vector-gDNA segments. Arrows indicate primers. Sequences are not drawn to scale. Photography of U. mutabilis was reprinted from Wichard and Oertel (2010) with permission from JohnWiley and Sons, Copyright© (2010) Wiley