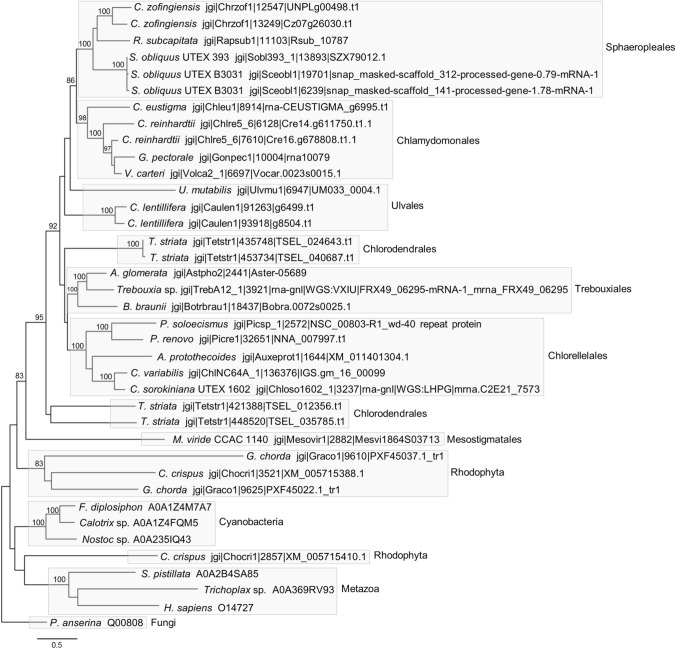
Fig. 4.
Maximum-likelihood phylogenetic tree using the LG + G + F substitution model based on the alignment of UM033_0004 and 37 identified homologs. Only the aligned APAF1_C/WD40 domain-containing regions were included (678 positions) with a gap cutoff of 85% (see Suppl. Data File 2). Bootstrap values (500 resamplings) > 80% are shown. The scale bar indicates the number of amino acid substitutions per site. Sequences are represented by the name of the organism from which they originated and the sequence identifier (see also Suppl. Data File S1). The NB-ARC domain-containing T. striata proteins have the identifiers TSEL_012356.t1 and TSEL_035785.t1