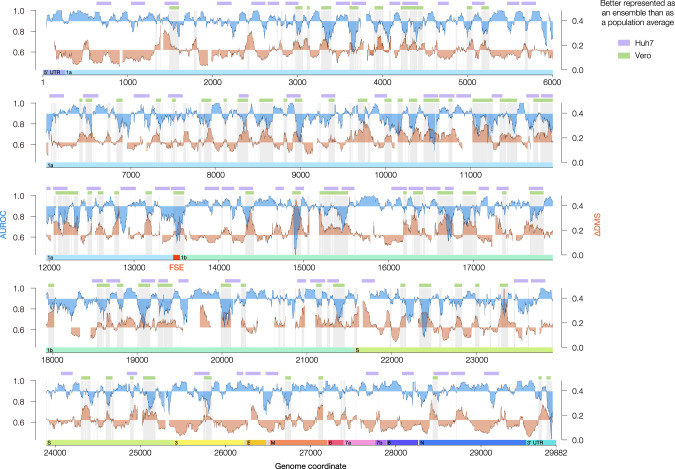
Fig. 3. Alternative RNA structures form across the SARS-CoV-2 genome.
Agreement between DMS reactivities and predicted secondary structures (AUROC, blue) and the difference in DMS reactivity between clusters 1 and 2 (∆DMS, orange) for the genome-wide model in Vero. Both quantities were calculated over sliding windows of 80 nt in 1 nt increments; x values represent the centers of the windows. Windows with <10 paired or <10 unpaired bases were excluded from the calculation of AUROC; windows with <10 bases that clustered into at least two structures were excluded from the calculation of ∆DMS. For AUROC and ∆DMS, the area between the local value and the genome-wide median is shaded. For the Vero model, all coordinates best described by structure ensembles (AUROC below median, ∆DMS above median) are shaded in light gray. The green bars represent a denoised version of these coordinates (see Methods). For the Huh7 model, regions meeting criteria for alternative structures (see Methods) are labeled with lavender bars. The locations of the untranslated regions (UTRs) and open reading frames (ORFs) of SARS-CoV-2 are indicated below the AUROC and ∆DMS data. The frameshifting stimulation element (FSE, coordinates 13,462–13,546) is highlighted in red. Source data are provided as a Source Data file.