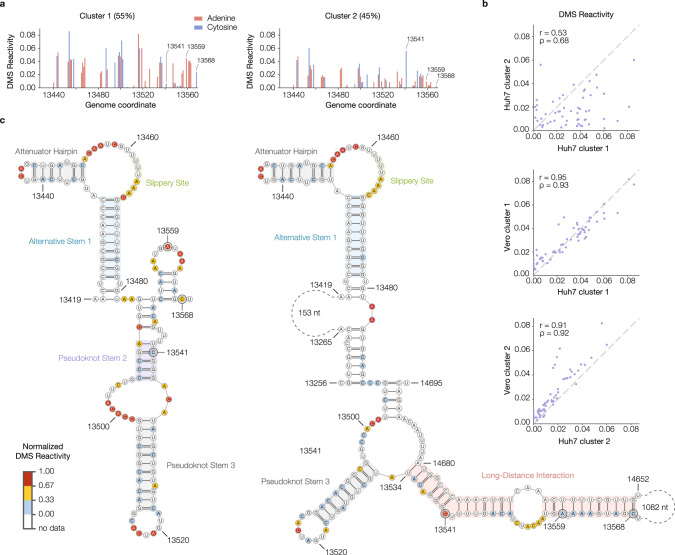
Fig. 5. Alternative conformations of the frameshifting stimulation element (FSE) derived from in-cell DMS-MaPseq data include a long-distance interaction.
a DMS reactivity profiles for both clusters from the Huh7 genome-wide RT-PCR data in the vicinity of the FSE (nucleotides 13,434–13,568). The abundance of each cluster is given beside its name. Each bar representing an adenine or cytosine is colored in red or blue, respectively. Three of the nucleotides whose reactivities differ substantially between clusters are labeled. b Scatterplots of DMS reactivities over the FSE, comparing the two clusters from Huh7 (top) and each Huh7 cluster with the corresponding cluster from Vero cells (middle and bottom). For each comparison, the Pearson (r) and Spearman (ρ) correlation coefficients are given. c Predicted structures of Huh7 clusters 1 and 2 based on DMS reactivities. In each structure, selected features are highlighted, including alternative stem 1 (in both clusters), a long-distance interaction (in cluster 2), and features that are also present in the canonical pseudoknot. The three nucleotides labeled in (a) are also labeled in the structure models. Nucleotides are colored by normalized DMS reactivities. Source data are provided as a Source Data file.