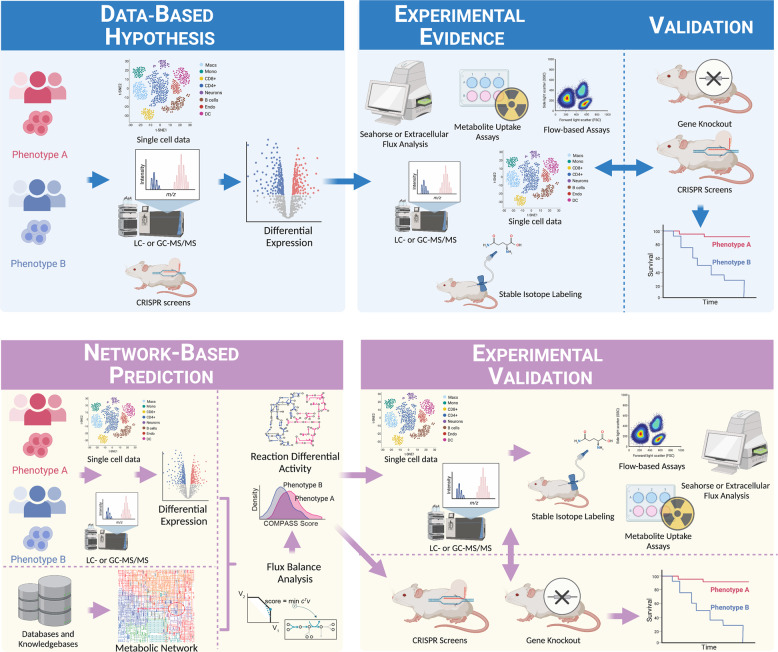
Fig. 1.
Network-based methods complement and add to enrichment-based workflows for interrogating immunometabolism. Top panel: High-throughput data, such as transcriptomics, metabolomics, or systemic CRISPR screens, are used to generate data-driven hypotheses in the form of differentially expressed targets [119, 120]. Pathways and gene sets are knowledge-based representations of shared biological activity derived from established gene ontologies and databases (e.g., GO [121, 122] and KEGG [61]). Subsequent experiments validate and refine the data-driven hypotheses, add mechanistic insight, and may lead to another cycle of high-throughput data collection and analysis. Bottom panel: Network-based approaches augment enrichment-based approaches. Biological networks, such as genome-wide metabolic networks, are generated from the annotated genome of a species of interest together with functional genomic data and computational gap-filling where appropriate [104–106]. Network algorithms, such as flux balance analysis for genome-scale metabolic models, integrate global information agnostic of pathway divisions and can therefore predict targets that will not be prioritized based on the workflow described above. Similar to pathway- and gene-set-based approaches, networks organize extant knowledge and allow the contextualization of gathered high-throughput data within extant knowledge bases. However, network approaches may capture systemic effects that are missed by pathway-focused approaches.