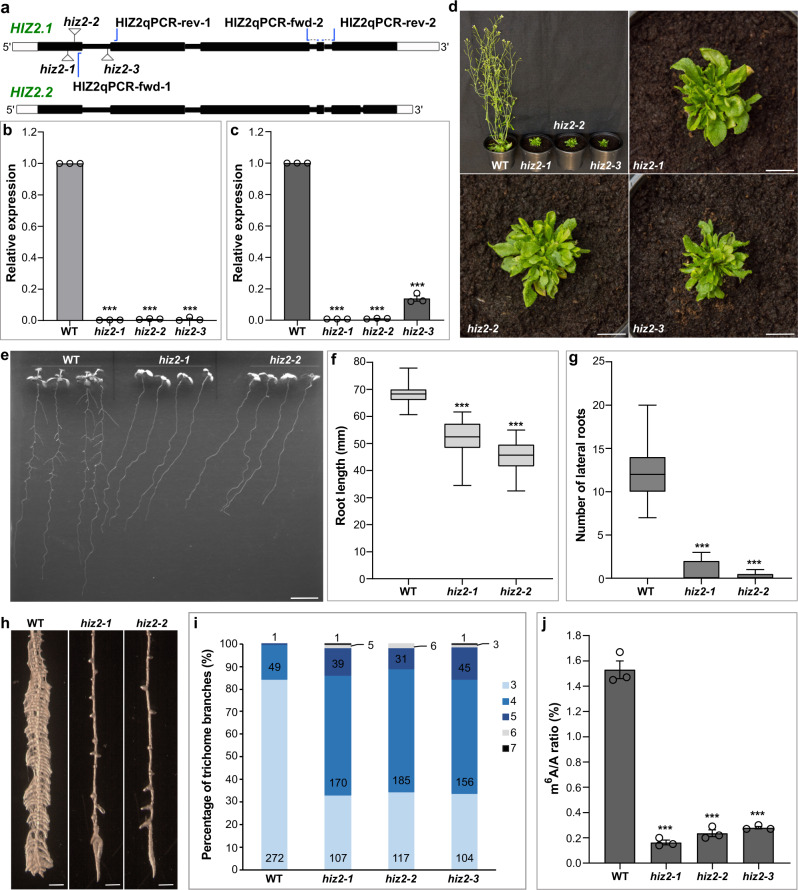
Fig. 6. Characterizing Arabidopsis HIZ2.
a Schematic of HIZ2 (AT5G53440) genomic DNA sequence with T-DNA insertion sites. HIZ2.1 and HIZ2.2 refer to two splice variants. White rectangles denote UTRs, black rectangles denote exons and thick black lines represent introns. hiz2-1 to hiz2-3 refer to Salk_120590, Salk_020625 and Salk_126486, respectively. HIZ2qPCR-fwd-1 and HIZ2qPCR-rev-1 were used as the first primer pair while HIZ2qPCR-fwd-2 and HIZ2qPCR-rev-2 were the second primer pair in RT-qPCR. b, c HIZ2 transcript levels checked by RT-qPCR using the first primer pair (b) and the second primer pair (c) labeled in a. CBP20 was used as a reference gene. Data represent mean ± SE from three biological replicates. Statistically significant differences relative to WT here and in f, g, j were analyzed by One-Way ANOVA (one-sided test) and marked with asterisks (***p < 0.001). d Five-week-old hiz2 mutants and WT planted in compost. Scale bar = 1 cm. e Eleven-day-old seedlings vertically cultured on 0.5× MS plus 1% sucrose. Scale bar = 1 cm. f, g Statistical data showing the root phenotypes of seedlings in e. Data represent mean ± SE (n = 25 individual seedlings). In box plots, the center line in each box indicates the median. The lower and upper bounds of each box represent the first quartile (25%) and the third quartile (75%), respectively. The bottom and top of whiskers denote the minimum and maximum, respectively. h Roots under the Stereo Dissecting Microscope after the treatment with N-1-naphthylphthalamic acid (NPA, 10 μM) and then transferred onto 0.5× MS with 1-naphthaleneacetic acid (NAA, 10 μM) for 5 days. Roots were soaked in 100% ethanol for a few seconds to minimize the effect of root hairs in photographing lateral roots. Scale bar = 500 μm. i Percentage of trichome branches on the first pair of rosette leaves from 2-week-old plants cultured in compost. Twenty leaves were used for counting the trichome branching in each line. j m6A levels checked by two-dimensional thin layer chromatography (TLC) analysis. Data represent mean ± SE from three biological replicates. Source data are provided as a Source Data file.