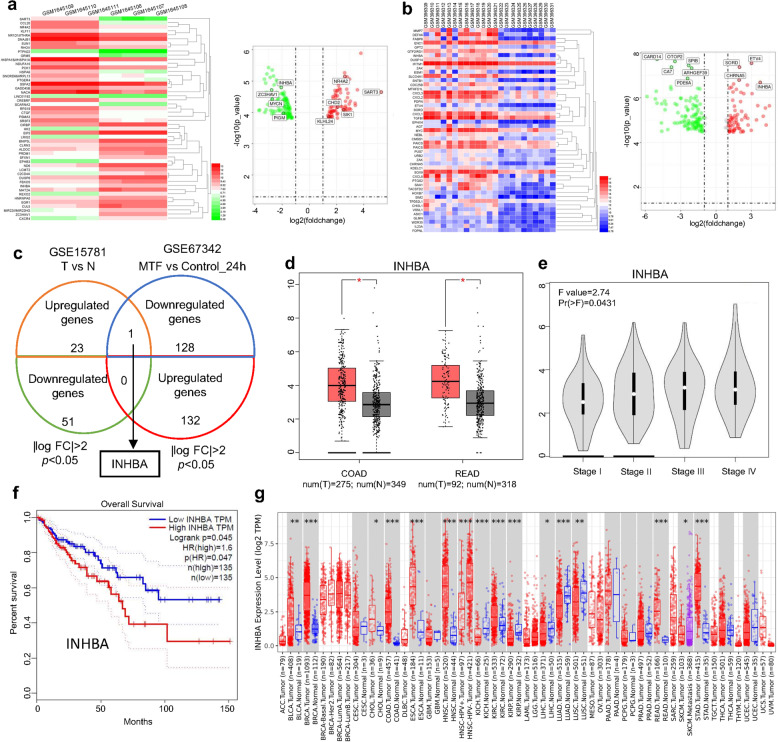
Fig. 2. Screening target gene of metformin in CRC.
a Analysis of the differential genes (DEGs) between metformin-treated group (24 h) and control group (24 h) in GSE67342 dataset. b Heatmap and volcano map showing DEGs between CRC tissues and adjacent tissues in GSE15781 profile. c Comparison of the genes altered in GSE67342 (|log2FC | >2; p < 0.05) with GSE15781 dataset (|log2FC | >2; p < 0.05). d, e GEPIA online database analyzing the mRNA expression of INHBA in CRC tissues and normal colon tissues (d), as well as the expression of INHBA in CRC tissues from the different stages (e). f Kaplan–Meier curve showing the OS probability of CRC patients from the GEPIA database with low or high mRNA levels of INHBA (log-rank test). OS overall survival. g Expression of INHBA in various tumor tissues and their adjacent tissues. Red boxplot: tumor tissues; Bule boxplot: normal tissues; Purple boxplot: metastatic tissues. The data was gained from the TIMER database.