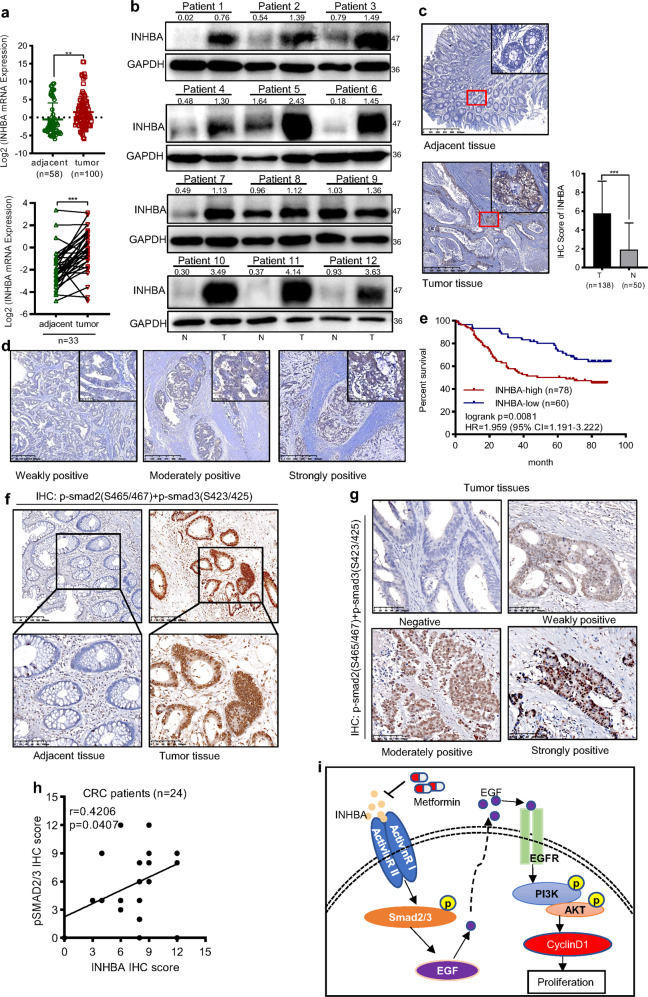
Fig. 8. INHBA was highly expressed in human CRC tissues and correlated with unfavorable prognosis.
a qPCR analysis of the mRNA levels of INHBA among CRC and adjacent normal tissues (Student’s t test), as well as 33 pairs of tumor tissues and adjacent normal tissues (paired t test) (low). b INHBA protein levels in fresh CRC (T) and adjacent normal samples (N) (n = 12) were detected by western blotting. c Representative IHC images of CRC tissues and corresponding normal tissues stained with an INHBA antibody (n(T) = 138, n(N) = 50) (Student’s t test). Magnification: 200× (small); 40× (large). Scale bar: 50 µm (small); 400 µm (large). d The representative immunohistochemical images showing different staining intensities of INHBA in CRC tissues. Magnification: 200× (small); 40× (large). Scale bar: 50 µm (small); 400 µm (large). e Kaplan–Meier curve showing the OS of patients with CRC according to INHBA expression (log-rank test). f Representative IHC images of p-smad2(S465/467) and p-smad3(S423/425) staining in CRC tissues and paracancerous tissues. g Representative images showing the staining strength of p-smad2/3 in CRC tissues. Magnification: 100×. Scale bar: 100 µm. h The correlation between INHBA expression and p-smad2/3 expression in 22 CRC tissues. i Schematic description of the main molecular mechanism of metformin suppressing CRC proliferation. OS overall survival, HR hazard ratio, CI confidence interval. The values are shown as the mean ± SD and experiments were independently repeated three times.