Figure 1.
Spatial transcriptomics to explore the tumor microenvironment of high-grade serous ovarian carcinoma (HGSC)
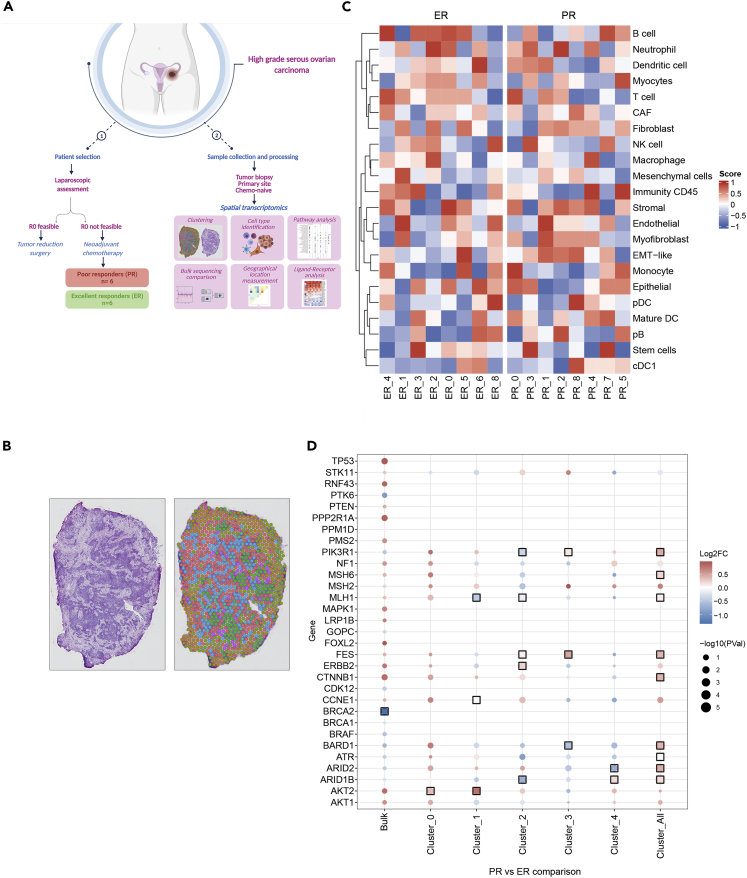
(A) Schematic representation of the study for tissue collection in patients with HGSC. R0, no gross residual disease. Created with BioRender.com.
(B) Representative image of a hematoxylin-and-eosin-stained section (top) and unsupervised clustering (bottom).
(C) Heatmap of putative cell composition of each cluster in the excellent responder (ER) and poor responder (PR) groups.
(D) Dot-plot representing differential gene expression of known ovarian cancer genes (COSMIC) in bulk RNA sequencing and in spatial transcriptomics of the main clusters (0–4). Black boxes are shown around the point when the gene set enrichment analysis adjusted p value is <0.1.