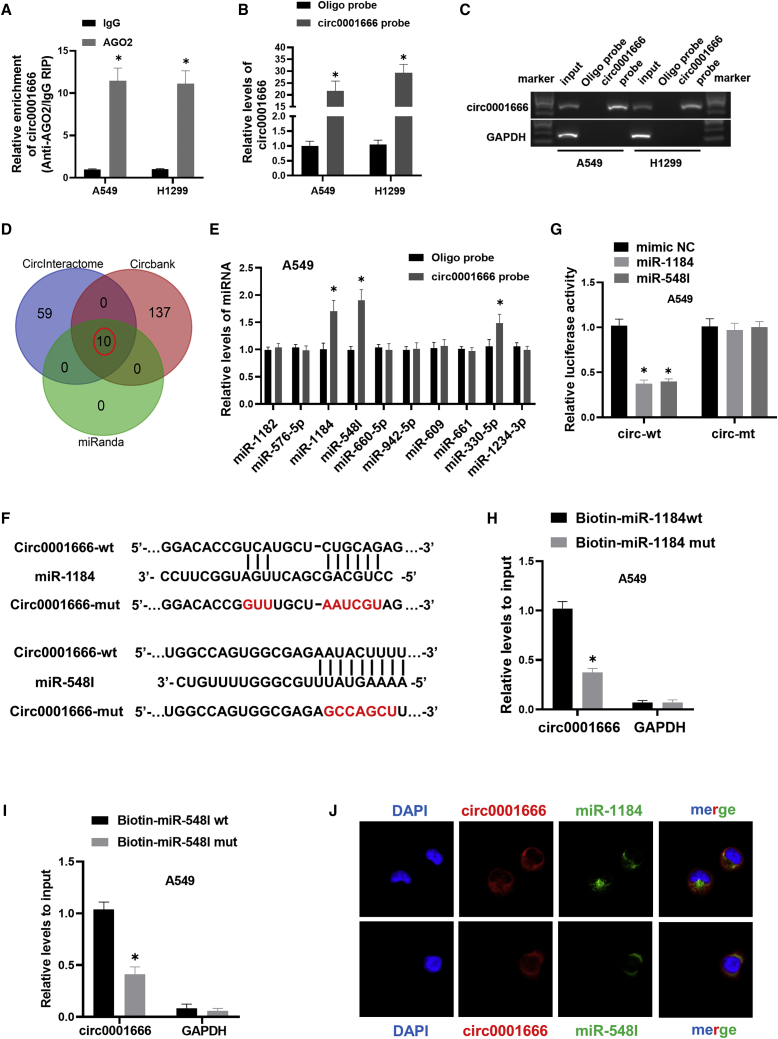
Figure 3.
circ0001666 could directly bind to miR-1184 and miR-548I in NSCLC cells
(A) Immunoprecipitation (RIP) assay was carried out using anti-AGO2 or IgG antibodies. circ0001666 levels in the samples were quantified using quantitative real-time PCR. (B and C) The efficiency of circ0001666 probe was measured by quantitative real-time PCR and gel electrophoresis. Relative level of circ0001666 was normalized to input. GAPDH was used as negative control. (D) Schematic illustration showed the overlapping target miRNAs of circ0001666 predicted by CircInteractome, miRanda, and Circbank. (E) The relative levels of 10 miRNAs were pulled down by circ0001666 probe or oligo probe in A549 cells. (F) Schematic graph showed the predicted binding sites between circ0001666 and miR-1184/miR-548I and mutation of potential miRNAs binding sequence in circ0001666. (G) The luciferase activities were detected in A549 cells after co-transfection with circ0001666-wt or circ0001666-mut and miR-1184/miR-548I mimics or mimics NC. (H and I) Relative expression levels of circ0001666 were captured by biotinylated wild-type miR-1184/miR-548I or mutant miR-1184/miR-548I. (J) FISH images showed the subcellular co-localization of circ0001666 and miR-1184/miR-548I. circ0001666 was stained red with Cy3, miR-1184/miR-548I were stained green with Dig, and the nuclei were stained blue by DAPI. ∗p < 0.05.