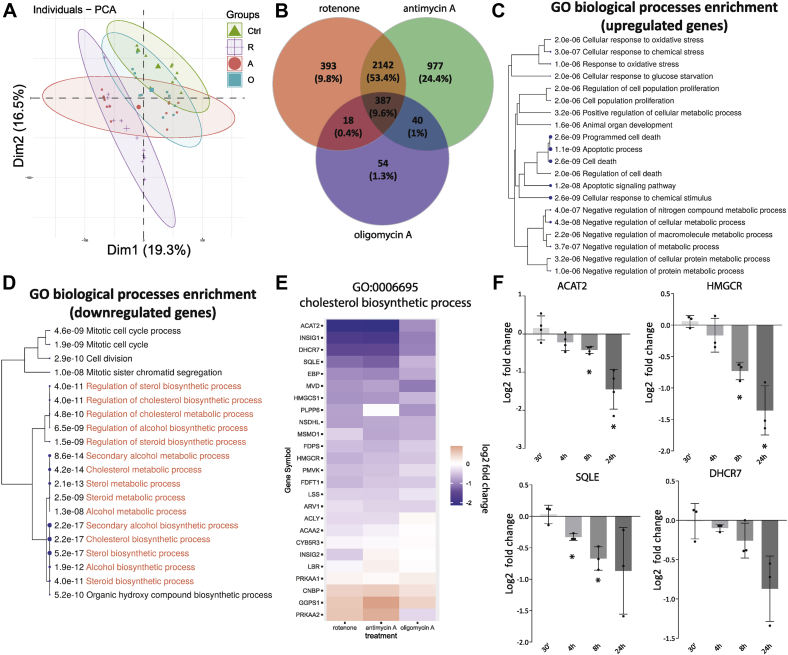
Figure 2.
Transcriptional adaptations shared in the three pharmacological models of respiratory chain dysfunction.A, PCA of QuantSeq data of human fibroblasts cultured for 1 or 5 days in the presence of respiratory chain inhibitors rotenone (100 nM; purple plus signs), antimycin A (500 nM; red circles), or oligomycin (1 μM; blue squares). Control values are from DMSO-treated cells (0.1%; green triangles). Separation along the first two dimensions is shown. Cells were plated to reach similar final confluency. B, Venn diagram illustrating the total number and overlap of the differentially expressed genes for the three treatment groups. C and D, biological processes associated with differentially expressed genes shared by all three respiratory chain inhibitors as shown in panel B (387 genes). A hierarchical clustering of the significantly enriched GO-biological process pathways is shown. For each pathway, the significance of its enrichment in the shared set of DEGs was calculated and shown in the figure. The processes most relevant for this study are highlighted in red of panel D. E, expression changes compared to DMSO (0.1%) control of genes associated with the cholesterol biosynthetic process (GO:0006695) following treatment with rotenone (left), antimycin A (middle), and oligomycin A (right). The heatmap representation reveals the consistent downregulation of most genes associated with this pathway. F, time-dependent changes in gene expression of five key mevalonate pathway genes in human fibroblasts measured by qPCR (see Experimental procedures). Cells were treated with rotenone (100 nM) for the indicated times. Data are shown as the log2-fold change of gene expression compared to control-treated human fibroblasts (0.1% DMSO). Bars show mean ± standard deviation; black circles indicate results from three independent experiments. Significance calculated as one-sample t test (H0: μ = 0). ∗ p< 0.05. DEG, differentially expressed genes; DMSO, dimethyl sulfoxide; GO, Gene Ontology.