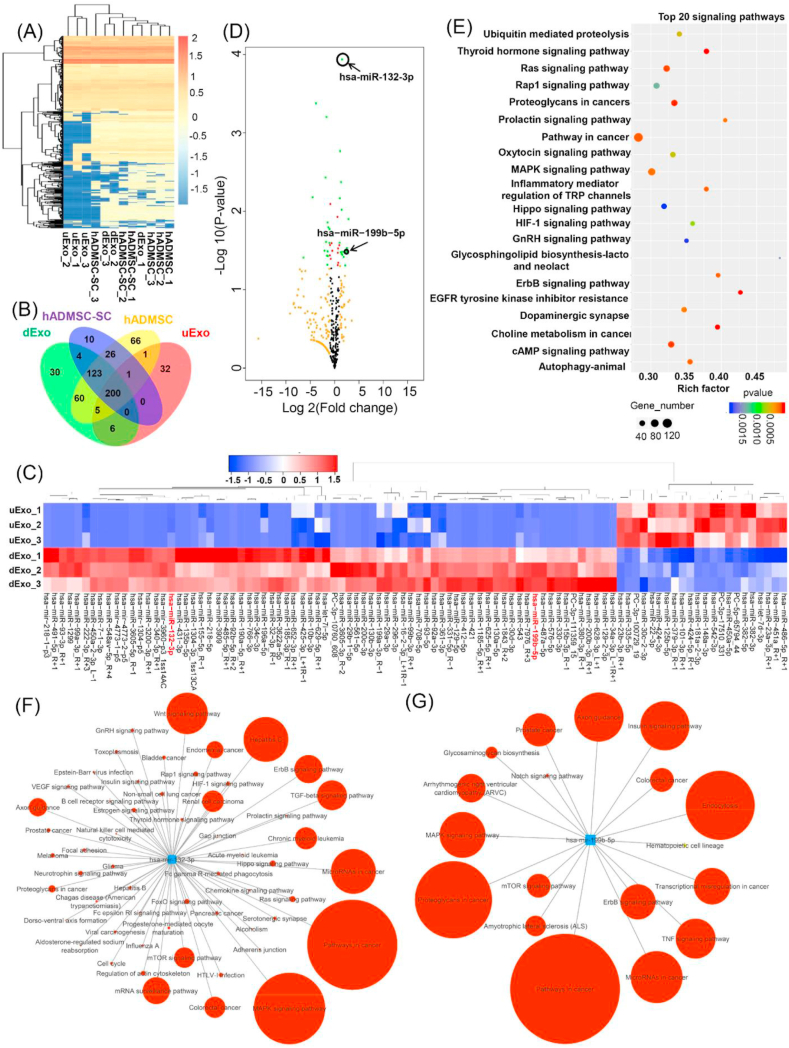
Fig. 7.
miRNA sequencing analysis. (A) Heatmap and hierarchical clustering depicting the normalized Z-scaled expression of miRNA in the four groups (n = 3 for each group). (B) The Venn diagram of all differentially expressed miRNAs identified in the four groups. (C) Heatmap of significant differential expression of miRNAs in uExo and dExo groups (n = 3, <p<0.05). (D) Volcano plot showing differences in exosomal miRNAs expressed in the uExo and dExo groups. Note for spot colors: black, insignificant; red, <p≤0.05; orange, log2FoldChange ≥1 or ≤ -1; green, both red and orange criteria fulfilled. (E) The top 20 common KEGG pathways of the differentially expressed miRNAs in the uExo and dExo groups. Number of target genes enriched in KEGG pathway, p value and rich factor are shown in scatterplot. Rich factor = (number of target genes in KEGG pathway)/(total number of genes in KEGG pathway). The networks predicted by the miRPathDB database for the upregulated miRNAs (miRNA-132-3p (F) and miRNA-199b-5p (G)), showing the associated functional signaling pathways. The circle size (larger to smaller) represents the number of hits or genes in the interacting pathway.