ABSTRACT
Fungi form a major and diverse component of most ecosystems on Earth. They are both micro and macroorganisms with high and varying functional diversity as well as great variation in dispersal modes. With our growing knowledge of microbial biogeography, it has become increasingly clear that fungal assembly patterns and processes differ from other microorganisms such as bacteria, but also from macroorganisms such as plants. The success of fungi as organisms and their influence on the environment lies in their ability to span multiple dimensions of time, space, and biological interactions, that is not rivalled by other organism groups. There is also growing evidence that fungi mediate links between different organisms and ecosystems, with the potential to affect the macroecology and evolution of those organisms. This suggests that fungal interactions are an ecological driving force, interconnecting different levels of biological and ecological organisation of their hosts, competitors, and antagonists with the environment and ecosystem functioning. Here we review these emerging lines of evidence by focusing on the dynamics of fungal interactions with other organism groups across various ecosystems. We conclude that the mediating role of fungi through their complex and dynamic ecological interactions underlie their importance and ubiquity across Earth's ecosystems.
Keywords: biotic interactions, fungal biogeography, symbiosis, mycovirus, mycobiota, omics
The unique set of traits that fungi exhibit, and the versatility of these traits facilitate the mediating role that fungi play in host and ecosystem functioning through the establishment of dynamic evolutionary and ecological interactions.
INTRODUCTION
Fungi are ubiquitous and form diverse communities temporally and spatially spanning multiple scales across many ecosystems (Fig. 1). We have only begun to scratch the surface of the importance of fungi in our lives and environment owing to their great diversity and challenges involved in studying fungal biology and ecology. Of an estimated 1.5–5 million fungal species only around 120 000 have been described (Hawksworth and Lücking 2017), largely due to the difficulty of isolating and culturing most fungi (Kõljalg et al. 2013), which has also been a barrier to assigning them functional roles. Furthermore, it is difficult to distinguish where an individual of a fungal species begins and ends. This is highlighted by the tight association of fungi with other organisms e.g. through lichen, mycorrhizal, endophytic and mixed biofilm associations, but also by potentially frequent cross-kingdom horizontal gene transfer (HGT) between fungi and other organisms (Venice et al. 2020b), and in the heterogeneity of genetic and phenotypic intraspecific and intracellular variation that occurs in fungi (Ehinger et al. 2012; Taylor et al. 2017). With recent advances in molecular methods for gene-based identification of microbes over the last 20 years, some of these challenges have been overcome, leading to a rapid growth in our understanding about the importance of fungi in various ecosystems (reviewed in Peay, Kennedy and Talbot 2016; Nilsson et al. 2019). In terrestrial habitats, while relying on their hosts for carbon resources, mutualistic and pathogenic fungi play key roles in driving the physicochemical and biological properties of their environment as well as host population dynamics (Alan Pounds et al. 2006; Clemmensen et al. 2013; Bagchi et al. 2014; Chen et al. 2019; Tedersoo, Bahram and Zobel 2020). Meanwhile saprotrophic fungi are the key decay agents of organic material, playing a central role in carbon and nutrient cycling in many ecosystems (Averill, Turner and Finzi 2014; Fernandez and Kennedy 2016; Netherway et al. 2021). In aquatic habitats, fungi can regulate food-web dynamics and biogeochemical cycling by rendering resources more available to higher order consumers and increasing the efficiency of trophic transfer through aquatic ecosystems (reviewed in Grossart et al. 2019).
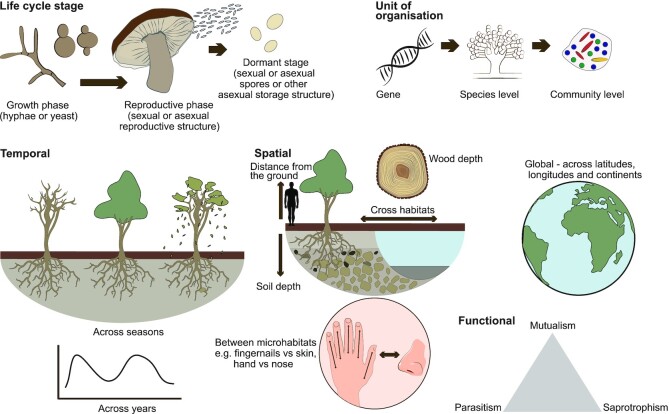
Figure 1.
Fungi form diverse entities that are distributed across different aspects and scales of space and time. Fungal diversity can be viewed across different levels of biological organization from the gene level to the level of complex communities, as well as from a taxonomic, phylogenetic or functional perspective. Even at the individual level, fungi exhibit different life cycle stages, that all have their unique biogeographical patterns e.g. the growth phase of fungi (yeast or filamentous) is embedded within or on a substrate, whereas the reproductive and dormant phases of fungi usually bridge different substrates/habitats. This is exemplified by a soil dwelling fungus from the family Agaricaceae that exists as a mycelium (growth phase) within the soil matrix, which then produces a fruitbody in the form of a mushroom that bridges habitats between the soil and the atmosphere. Finally, the fungus releases spores that may make their way into the atmosphere before landing in a new patch of soil, each growth phase is subjected to unique environmental factors, biological interactions, and other processes that lead to distinct biogeographic patterns. As fungi bridge the gap between micro- and macro-organisms, they also exhibit spatial patterns at the microhabitat to macro habitat level, including across different depths of a substrate, between different substrates, between different organs of a host, through to macroecological scales such as across landscapes to biomes and the global level. Fungi also exhibit distinct biogeographic patterns across different temporal scales from days to seasons, through to years and geologic time.
With large and varying phylogenetic and functional diversity as well as great variation in dispersal modes, fungi can act as important pathogens, commensals, and mutualists of macro-eukaryotic hosts, and interact with and influence the activity and functions of microbial prokaryotes and eukaryotes in most ecosystems (Fig. 2). They bridge the divide between micro- and macroorganisms, from single celled yeasts to complex macro-fungi that produce structures rivalling plants and animals in terms of size. Recent studies have provided unprecedented insights into the ecology and biogeography of fungi. There is growing evidence pointing towards commonalities and differences in the community assembly patterns and processes of fungi compared to those traditionally considered for microorganisms such as bacteria, but also from macroorganisms such as plants and animals (Tedersoo et al. 2014; Davison et al. 2015; Tisthammer, Cobian and Amend 2016; Kivlin et al. 2017; Bahram et al. 2018; Cameron et al. 2019). The characteristics of fungi and their ecology, including their associations with other organisms, may have enabled fungi to become central agents of ecosystem, ecological, and evolutionary processes (Fig. 2), and to span multiple scales and dimensions of time, space, and biological interactions (Figs 1 and 3). The combined and increasing evidence suggests that fungi can mediate links between different organisms and ecosystems, and potentially alter ecological, evolutionary and biogeographic relationships of the organisms they interact with. Here we review this evidence by exploring the current knowledge around the dynamics of fungal ecological interactions and underlying mechanisms in various ecosystems and discuss how these affect the evolution, ecological fitness and distribution of their associated organisms. We further present a perspective for embracing the importance of fungal interactions in ecosystems.
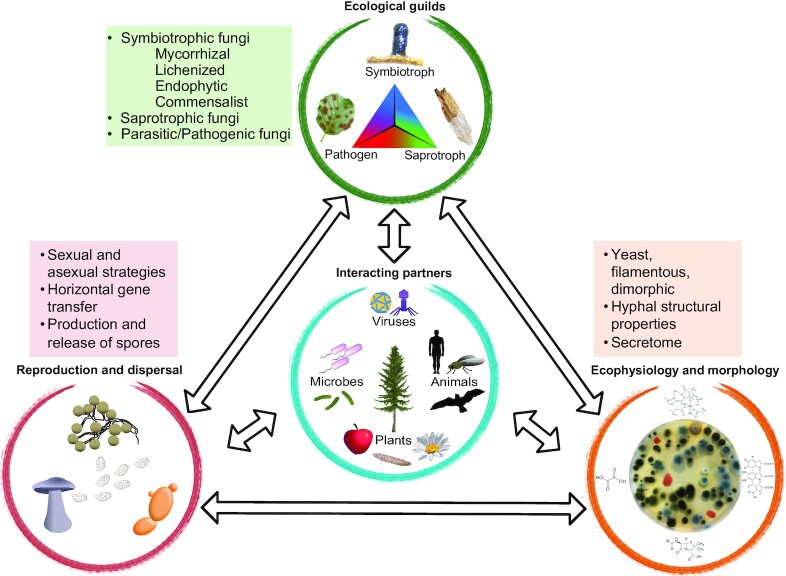
Figure 2.
Proposed triangle of fungal traits that are integral to their success as organisms and as links between organisms and across ecosystems. Fungal reproductive and dispersal traits are central to adaptation and dispersal to new environments, whereas fungal ecophysiological and morphological traits are central to their nutrient acquisition, stress tolerance, and interactions with other organisms. These two groups of traits then feed into the major fungal ecological strategies that transition along a three-dimensional continuum from symbiotrophism, saprotrophism to parasitism depending on environmental conditions. All groups of traits both influence and are influenced by interactions with other organisms, and these collections of traits allow fungi to be key modulators of ecological, ecosystem and evolutionary processes.
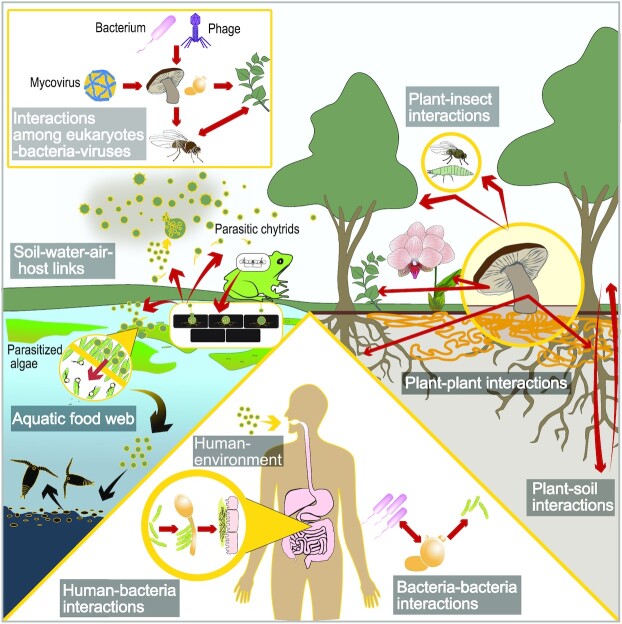
Figure 3.
Examples of mediating role of fungi in different ecosystems. Fungi through their unique morphological and ecophysiological properties act as mutualists, commensalists, and antagonists with plants, animals, microbes and other fungi, mediate the health, performance, population dynamics and biogeography of these organisms. Meanwhile, through their affinity for and ability to break down complex substrates (notably plant derived) and even contribute to mineral weathering, fungi mediate carbon and nutrient cycles in both terrestrial and aquatic ecosystems, and through their enormous production and release of spores into the atmosphere fungi may even mediate rainfall. For example, in terrestrial systems mycorrhizal fungi mediate nutrient acquisition of plants as well as their interactions with antagonists such as plant pathogens and herbivores, while saprotrophic fungi mediate the cycling of complex plant derived substrates, and fungal pathogens mediate the population dynamics of eukaryotic hosts. In aquatic systems fungi mediate food web dynamics by controlling resource fluxes to higher order consumers in the process also mediating the efficiency of transfer across trophic levels. In human habitats such as skin and gut, the mycobiome, through complex interactions with other microbes, can play a key mediating role in human health and dysbiosis.
THE ECOLOGICAL VERSATILITY OF FUNGI
Fungi exhibit diverse lifestyles (nutritional modes), reproductive and dispersal strategies and physicochemical traits, which could play important roles in their success (Fig. 2). Nutrient, carbon, and water acquisition is the central function of all organisms, and the heterogeneous distribution of these resources spatially and temporally is the driving force of evolutionary diversification and adaptation. Fungal hyphae with their indefinite growth habit and the formation of highly interconnected mycelial networks represent a key evolutionary adaptation to terrestrial life that facilitates the foraging of unevenly distributed resources and maximizing the efficient assimilation of these resources between distal patches with incredible plasticity (Fricker et al. 2017; Kiss et al. 2019).
Ecophysiological traits
Recent work suggests that we have likely just scratched the surface in terms of understanding the ecophysiological capabilities of fungi. The well-developed and diverse secretome (i.e. the total set of molecules secreted by cells) of fungi contributes to their metabolic versatility by establishing saprotrophic, mutualistic and pathogenic interactions (Bouws, Wattenberg and Zorn 2008; Choi et al. 2010; Girard et al. 2013). Fungal secretomes overlap greatly between mutualistic, saprotrophic and pathogenic lifestyles (Plett and Martin 2015; Hess et al. 2018), thereby potentially facilitating lifestyle transitions in order to evolve and adapt to varying environmental conditions (Girard et al. 2013). These secretomes enable fungi to extract nutrients from differing and complex substrates, but also protect fungi from hostile environmental conditions (Girard et al. 2013). Symbiotic and free-living fungi can inhabit extreme but vast areas of drylands and play an important role in rock weathering and thereby soil formation and create microhabitats for other organism groups (Coleine et al. 2021). Combined with their hyphal forming habit, perhaps as their key characteristics, the enzymatic arsenal of fungi has facilitated their dominance in inhabiting and decomposing plant derived substrates. The high diversity of carbohydrate-active enzymes (CAZymes) of pathogenic and saprotrophic fungi gives them the flexibility to adapt to different plant species and their resources, and the diversification of fungal nutritional modes is evolutionarily linked to the diversification of angiosperms and gymnosperms (Janusz et al. 2017). A recent study suggests that fungal species with the highest capacity and broadest diversity of enzyme substrates have unexpected lifeforms, such as dark septate endophytes and endolithic black yeasts, while those species traditionally exploited for industrial purposes rank quite low in terms of such enzymatic parameters (Lange, Barrett and Meyer 2021).
The diversity of secondary metabolites produced by fungi, which are central to both offensive and defensive responses (Rohlfs and Churchill 2011), facilitate associations with hosts (Keller 2019), while (positively or negatively) interacting with and influencing bacteria in establishing interactions with hosts (Bahram et al. 2018; Durán et al. 2018; Pierce et al. 2021). In addition, fungal secondary metabolites play a role in spore germination, either inhibiting spores from competing species or promoting their own (Becker et al. 2012; Khalid et al. 2018). Though underexplored, at least some key fungal secondary metabolites—including aflatoxins, melanin and β-lactam antibiotics—appear to be highly compartmentalized in vesicles that aid in their storage and transport under various conditions such as light and nitrogen limitation (Chanda et al. 2009; Upadhyay et al. 2016; Martin 2020). Fungi can regulate their secondary metabolite production based on environmental conditions, in particular light and temperature via both transcriptional and epigenetic regulations (Keller 2019).
As another important trait in lifestyle flexibility, fungal melanisation (which occurs in most fungal clades) allows fungi to resist environmental stressors (such as high temperatures, free radicals) and offers photoprotection (Cordero and Casadevall 2017), such as against UV radiation. Fungal melanin can also act as a virulence factor (reviewed in Jacobson 2000) and plays an important function in fungal infection across diverse hosts, supported by evidence that highly melanized fungi (belonging mainly to the Dothideomycetes class) are more capable of infecting organisms across different kingdoms, and switching and sharing hosts including between plants and humans (Gauthier and Keller 2013) as well as having broad geographical distributions. There have been several human disease outbreaks caused by highly melanized fungi which have likely been directly sourced from the environment, and it appears that the breakdown of physical barriers protecting against direct infection of fungi and their abilities to utilize host nutrients play an important role in the onset of fungal outbreaks (reviewed in Gauthier and Keller 2013). A recent example of an outbreak caused by a highly melanized fungi is the ‘black fungus’ epidemic infecting Covid-19 patients with weakened immune systems in India (Dyer 2021). While it remains unclear to what extent melanin contributes to fungal virulence, overall melanized fungi appear more capable to resist host defences compared to non-melanized fungi.
Lifestyle versatility
Another striking property of some fungi is their ability to transition between different lifestyles, which has enabled them to associate with a wide variety of hosts in various environmental conditions and adapt to spatially and temporally heterogeneous resources. While fungi have evolved different lifestyles, certain groups have retained many of the ancestral genes that enable transition between different nutritional modes (Hess et al. 2018). This capability together with retaining genomic potential to associate with a wide variety of hosts while adapting to new conditions (Genre et al. 2020), facilitated by HGT events (Wang et al. 2021) or by exchange of viruses via horizontal viral transfer (HVT) (Márquez et al. 2007), could contribute to host range expansion of mutualistic fungi, which is reflected in the relatively low host specificity of mutualistic in contrast to pathogenic fungi (Põlme et al. 2018). The frequent HGT and HVT events between fungi and their hosts (Bian et al. 2020; Wang et al. 2021) may also have enhanced their adaptive capacity with their hosts, including terrestrialization (Wang et al. 2021). A recent study provides direct evidence that fungal viruses can contribute to fungal lifestyle versatility by mediating transitions from pathogenic to non-pathogenic phenotypes (Zhou et al. 2021). There is also evidence that HGT events occur more frequently between different fungal species, compared to other eukaryotic groups (Khaldi et al. 2008), which can further contribute to their ecological versatility.
A particularly interesting case of evolutionary fungal lifestyle transitions is represented by the ectomycorrhizal (EcM) lifestyle, a convergent lifestyle that has arisen independently around 80 times or more from ecologically diverse saprotrophic ancestors (Miyauchi et al. 2020). While not true EcM fungi, a diverse variety of saprotrophic fungi can facultatively colonize the roots of plants; however, the functional significance of this relationship is unknown, though it perhaps represents a transitional stage between saprotrophy and an EcM symbiosis (Smith et al. 2017). This convergent evolution is in stark contrast to the arbuscular mycorrhizal (AM) symbiosis which has arisen once and remained relatively unchanged over hundreds of millions of years, although the AM symbiosis can perhaps act along a mutualism-parasitism continuum depending on host and environmental conditions (Johnson et al. 2015). The versatile boundaries between fungal lifestyles appears to be the norm rather than the exception. One piece of evidence supporting this view is that most of the fungi capable of infecting both humans and plants are soil saprotrophs that can be opportunistic pathogens (Gauthier and Keller 2013). Dimorphic switching between a filamentous form and yeast form is a particularly powerful trait that facilitates the transition between lifestyles based on environmental conditions and habitats. This is exemplified by thermally dimorphic fungal pathogens that have evolved the ability to switch from a hyphal form in the environment to a single celled yeast form when infecting an animal host, a phenomenon that is triggered by elevated temperatures (Li and Nielsen 2017). Such versatility in switching between various symbiotic and free-living lifestyles may give fungi a unique capability to survive under unfavourable host and environmental conditions.
Reproductive traits
Reproduction is at the centre of life, with both sexual and asexual reproduction incurring both costs and benefits. Fungi appear to exhibit diverse and often unique and cryptic modes of reproduction, ranging from asexual to sexual, and often both strategies are employed to some degree (Nieuwenhuis and James 2016). It is often assumed that sexual recombination should be most advantageous in terms of accelerating evolutionary adaptation through natural selection and eliminating deleterious mutations (Aanen and Hoekstra 2007). Nevertheless, around 20% of fungal species may be solely asexual (Dyer and Kück 2017), including many evolutionary and ecologically successful fungi such as members of the Glomeromycota that form AM associations, although the presence of strictly asexual reproduction in the Golmeromycota is debated with growing evidence for the contrary (Yildirir et al. 2020). In addition, a sexual cycle has recently been observed in other fungal species previously thought to be solely asexual (Dyer and Kück 2017). Interestingly several fungi exhibit an alternative mechanism to sexual recombination in the form of parasexual recombination (Pontecorvo 1946), which is unique to the fungal kingdom and facilitates genetic transfer without meiosis and the production of sexual structures (Steensels, Gallone and Verstrepen 2021). Parasexuality in fungi may play a key role in pathogenesis in both plant and animal hosts (Calo, Billmyre and Heitman 2013), exemplified by Candida albicans in humans, and the rice blast fungus Magnaporthe oryzae in plants (Steensels, Gallone and Verstrepen 2021). Thus, the diversity and flexibility of reproductive strategies in fungi appears to play a central role in their evolutionary and ecological success. These reproductive strategies play a central role in the adaptation and dispersal of fungi to new environments, as discussed below.
Dispersal traits
Together with their reproduction strategies, the unique dispersal traits of fungi play a key role in their ability to span multiple scales of space, time and biological interactions (Fig. 1). Sporulation is a successful and phylogenetically widespread strategy for survival and dispersal that is employed by species of bacteria, protists, fungi and plants, yet fungi may represent most of the spore-forming organisms of the biosphere (Huang and Hull 2017). Sporulation contributes to the ease of fungal dispersal via different vectors. One of these vectors is air currents, which can carry a large proportion of biogenic aerosols comprising fungal spores, as evidenced by a high relative abundance and diversity of fungi in air (Gusareva et al. 2019). Another important dispersal vector for fungi is animals, which range from passive to tightly evolved symbioses and aid in both short and long-distance dispersal (Golan and Pringle 2017; Nuske et al. 2019; Vašutová et al. 2019). Animal associated fungi that are adapted to their host environment may show limited distribution following their hosts, such as fungus-growing ants and termites with limited distribution ranges (Biedermann and Vega 2020). Another example is the association between truffle-like fungal taxa which—due to forming underground fruiting bodies—rely on mycophagous mammals (Nuske et al. 2019) and birds (Caiafa et al. 2021) for dispersal, while the distribution of fungus-dependent animals can be heavily influenced by the production and distribution of fungal fruiting bodies (Claridge 2002). Also, some fungal individuals such as members of the genus Armillaria can disperse over large distances via vegetative growth, forming extensive mycelial networks that may form some of the largest organisms on Earth (Anderson et al. 2018). Overall, there appears to be large variation in the reproductive and dispersal abilities of different fungal taxa that may enable them to occupy complementary niches across various ecosystems.
Genomic traits
Last, but not least and connected in some respect to all the above-mentioned traits, one of the main features of fungi is their relatively small and dynamic genomes, which could lead to their greater ecological plasticity compared to other eukaryotes. This is supported by recent evidence suggesting that among AM fungi, those with large genomes show less versatility in host and habitat preferences, as shown in the case of Gigaspora margarita possessing by far the largest annotated fungal genome (Venice et al. 2020b). Also, parasitic fungi associate with a broader range of host species, compared to animal or protozoan parasites with much larger genomes compared to fungi (Ogburn 2019). In addition, the large variability of genome size among fungi can be mostly ascribed to the abundance of transposable elements, which in turn has been shown to be related to the evolution of symbiosis as well as the versatility of fungal lifestyles and genomic plasticity (Hess et al. 2014; Castanera et al. 2016; Faino et al. 2016; Frantzeskakis et al. 2018; Miyauchi et al. 2020). This is thought to be of particular importance for plant pathogenic fungi in agricultural landscapes that can support large and stable populations of pathogens that can rapidly adapt and evolve new virulence traits in response to host resistance factors even with low effective population sizes, in part due to the accumulation of and variability in the presence of transposable elements within species and between closely related species (Möller and Stukenbrock 2017). The adaptive evolution of fungi in their host environment may extend beyond their genomes, as evidenced by a recent study pointing to the role of epigenetic factors in plant and human fungal pathogens in developing resistance against antifungal agents (Torres-Garcia et al. 2020). Fungal secondary metabolism can also be highly flexible and manipulated by epigenetic chromatin regulators (Pfannenstiel and Keller 2019); notably methylation patterns in fungi appear to be maintained by natural selection not acting on the DNA sequence as the substrate, hinting at the inheritance of phenotypic traits (Madhani 2021).
Taken together, the ecological versatility of fungi seems unrivalled by other organism groups, which is facilitated by their unique dispersal, genomic, morphological, and ecophysiological attributes that are often plastic, thus considering them within a larger framework of traits including biotic interactions (Fig. 2), provides a more integrated understanding of fungal ecology and their central role in ecosystem and host functioning.
EVOLUTIONARY AND BIOGEOGRAPHICAL PERSPECTIVES
The early diverging fungi appeared to be endosymbiotic of eukaryotic hosts (Bonneville et al. 2020) and relied on their hosts for primary metabolism (Berbee, James and Strullu-Derrien 2017; Strullu-Derrien et al. 2018). These early fungi were zoosporic, which diverged more than 700 million years ago into non-flagellated fungi, followed by the most diverse fungal lineage of Dikarya around 642 Ma (Tedersoo et al. 2018). These diversification events were associated with fungal land colonization, which coincided with the diversification and colonization of land by plants and animals (Berbee, James and Strullu-Derrien 2017; Minter et al. 2017; Lutzoni et al. 2018).
Plant-associated fungi
It has been hypothesized that mycorrhizal-like fungi, may have facilitated the successful colonization of land by plants through acting as their primary nutrient uptake mechanism in a nutrient-limited environment (Pirozynski and Malloch 1975), which is plausible given the wide range of extant plants associating with mycorrhizal fungi (>90% of plant families). A growing body of evidence supports this hypothesis. For example, fossil records show the emergence of fungi prior to plants as well as the presence of arbuscular mycorrhizal (AM) fungus-like remnants in embryophytes (i.e. earliest diverging land plants (Remy et al. 1994; Redecker 2000; Bonneville et al. 2020; Gan et al. 2021), while phylogenetic analyses show the diversification of terrestrial fungi predates that of embryophytes (Lutzoni et al. 2018). Furthermore, experimental results show orthologous genetic pathways are involved in the symbiosis of AM fungi with embryophytes and vascular plants (Rich et al. 2021) and CO2-rich conditions during early plant evolution likely enhanced the net benefit of the AM symbiosis (Humphrey et al. 2018). Recent studies also provide evidence that the evolution of mycorrhizal symbioses has led to the diversification of EcM fungi and their host plants, based on comparing the centre of diversification and origin of hosts with their fungal partners (Looney et al. 2016; Brundrett and Tedersoo 2018). A few studies based on molecular phylogenies show that while some clades may originate from the tropics (e.g. Clavulina (Kennedy et al. 2012)), several major EcM clades appear to show a higher diversification rate in temperate regions (Kennedy et al. 2012; Looney et al. 2016). The diversification centres of at least some EcM fungal clades may have coincided with the distribution range expansion of their host plants, which is greater in temperate regions (Steidinger et al. 2019). This is further supported by shared evolutionary histories of plants and EcM fungi like those belonging to the genus Amanita (Sánchez-Ramírez et al. 2015). In terms of contemporary plant biogeography, due to their heavy reliance on fungal partners in their earlier life stages, the distribution ranges of orchids can be limited by the presence of suitable mycorrhizal fungi (McCormick, Whigham and Canchani-Viruet 2018). There is also evidence suggesting that the limited distribution of mycorrhizal trees beyond the tree line might be at least partly due to a lack of suitable fungal partners (Shemesh et al. 2020). Plant-associated fungi such as mycorrhizal fungi clearly play a central role the in macroecology of plant communities owing to their tightly intertwined evolutionary histories (Tedersoo, Bahram and Zobel 2020).
Animal-associated fungi
Much less is known about the evolution of animal-associated fungi, despite being phylogenetically more closely related, compared to fungi and plants (Wainright et al. 1993). Based on fossil evidence, the earliest animal associated fungi are likely parasites that shifted their interactions from plants to insects (Lepidoptera), and these fungi show a Cretaceous diversification like that of their insect and plant hosts (Sung, Poinar and Spatafora 2008). There is also molecular phylogenetic evidence that an entomopathogenic fungus (Ophiocordyceps unilateralis) specific to formicine ant species might have co-diversified with its host (Kobmoo et al. 2012), and these associations date back to the mid Paleogene based on fossil records of ant feeding behaviour driven by the fungus (Hughes, Wappler and Labandeira 2011). This is supported by phylogeographic analysis of a closely related species (O. sinensis) and its host insects, which showed that both interacting partners have influenced the evolution and diversification of each other (Zhang et al. 2014).
Biogeographic patterns of host-fungal interactions
Large-scale distribution patterns of fungi also support co-diversifications and similar evolutionary histories of symbiotic fungi and their hosts. Many fungal groups show similar diversity distribution patterns as their hosts. In particular, the diversity of animals and fungal animal parasites and pathogens follow a latitudinal diversity gradient, i.e. they are highest in tropical regions relative to other biomes (Willig, Kaufman and Stevens 2003; Tedersoo et al. 2014), suggesting a potential causal link between diversity of parasites and animal hosts. Plant-associated fungi, such as foliar endophytes (Arnold and Lutzoni 2007), AM fungi (Davison et al. 2015) and plant pathogens (Tedersoo et al. 2014), appear to also show an increasing diversity pattern towards the equator (Arnold and Lutzoni 2007; Tedersoo et al. 2014; Davison et al. 2015), which follows that of their host plants (Francis and Currie 2003; Hillebrand 2004; Kreft and Jetz 2007). Nevertheless, compared to other groups of symbiotic fungi, EcM fungi also exhibit a contrasting latitudinal diversity gradient, being highest in temperate and boreal forests compared to tropical forests (Tedersoo et al. 2012). This has been ascribed to the greater cover of EcM trees in temperate and boreal forests compared with tropical forests, which is consistent with the species-area relationship, as also reflected in higher species diversification rates of different EcM fungal clades (Kennedy et al. 2012; Tedersoo et al. 2012).
Multipartite evolutionary dynamics
In addition to their historical interactions with animals and plants, fungi have likely been exposed to gene exchanges with bacteria through HGT events—because of their shared niches and constant physical contact—and this may have enabled fungi to more efficiently use carbon sources and interact with other eukaryotes including plants (Naumann, Schüßler and Bonfante 2010; Bruto et al. 2014; Mares et al. 2015; Torres-Cortés et al. 2015; Venice et al. 2020b). Such tripartite interactions may be ancestral, as shown in the case of the AM symbiosis (Naumann, Schüßler and Bonfante 2010). A recent study suggests that by strengthening each other's fitness (i.e. fitness alignment), interactions of AM fungi and bacteria (particularly nitrogen-fixing) with the same host plant may synergistically shape the evolutionary trajectory of the three interacting partners (Afkhami, Friesen and Stinchcombe 2021). Although the underlying mechanism remains little explored, it is likely that HGT events among these mutualistic partners contribute to their fitness alignment, as implied by evidence of cross-kingdom gene transfer among AM fungi, bacteria, and plants (Li et al. 2018; Venice et al. 2020a,b). Besides increased adaptations to new soil environments and hosts as a result of HGT events, fungi may have also facilitated the transfer of bacterial genes—in addition to those from fungi—to plants, with potential implications for plant-fungal co-evolution and their co-diversification (Sinn and Barrett 2020). This is plausible given that fungi could be intermediate links between bacteria and plants, which is supported by evidence for ancient HGT events between mycorrhizal fungi with both bacteria and plants (Venice et al. 2020b). It remains to be seen whether HGT events have been more frequent between plants and their specific symbiotic fungi, e.g. between EcM plants and fungi. Fungal hyphae are also known to act as an anchor for attachment and colonization of bacteria inside plant tissues, as well as the formation of mixed biofilms that act as HGT hotspots and enable microbes to better cope with unfavourable conditions on host plants (reviewed in Overbeek and Saikkonen 2016).
Another interesting evolutionary and ecological interaction between fungi and their eukaryotic hosts comes in the form of viruses. Fungi and their viruses can contribute to plant adaptation to extreme environmental conditions. This has been shown in the case of a tropical panic grass, where a fungal endophyte confers heat resistance to the plant but only when the fungus is infected with a specific virus, suggesting a tripartite plant-fungus-virus symbiosis (Márquez et al. 2007). Fungal viruses may also play a key role in determining the expression of various traits of endophytic and phytopathogenic fungi including growth rate, morphology and pathogenicity (Xie and Jiang 2014; Zhou et al. 2021). Viruses are common in both plants and fungi, and—as eukaryotes—fungi and plants share viral families with closely related taxa that are genomically integrated in both hosts (Liu et al. 2010). Yet, the same does not appear to be as common between fungi and animals, which are more closely related than fungi and plants, once again highlighting the strong evolutionary and ecological link between fungi and plants since splitting from animals (Roossinck 2019). Thus, HGT events via shared or closely related viruses are perhaps a key mechanism that facilitates the intimate relationships fungi have with plants as endophytes, pathogens and mycorrhizal fungi. In support of this hypothesis, there is a growing number of studies pointing to compatibility and cross-kingdom HVT between fungi and plants (Andika et al. 2017; Nerva et al. 2017; Bian et al. 2020; Sutela et al. 2020), which could have contributed to the evolution of their viromes. Evidence also suggests that fungi can facilitate the spread of plant viruses to other plant hosts, e.g. via fungal spores as a vector (Bian et al. 2020). Viruses may exploit mycophagous insects to transmit between plant-pathogenic fungal hosts and form a complex quadripartite virus-fungus-insect-plant interaction with ecological and evolutionary implications (Liu et al. 2016). How exactly fungi and viruses co-evolve remains to be established, but it is plausible that their interactions contribute to fungal success in many ecosystems as discussed in the following sections.
Overall, evidence points to the role of fungi in the evolutionary ecology of their hosts and suggest that land colonization might have been a breakpoint in fungal diversification and evolutionary trajectories, owing to the formation of tight associations and interactions with other organism groups (Naranjo-Ortiz and Gabaldón 2019).
FUNGAL INTERACTIONS IN HOST SYSTEMS
As fungi are a major component of the microbiota in many ecosystems (Bahram et al. 2021), their interactions with hosts and other microbes are extremely relevant to consider in understanding host and ecosystem functioning. Although the underlying mechanisms remain unclear, there is a growing body of knowledge pointing to the importance and dynamics of fungal interactions and the role of fungi in mediating interactions between other organisms across ecosystems as discussed below (Fig. 3).
Fungal-plant interactions
The association between plants and fungi is one of the most well studied interactions. Plants as sessile organisms have evolved to live in close association with diverse communities of microbes- of which fungi are a major component—in order to meet their nutritional requirements for growth, survival and reproduction. Thus, they have developed multiple strategies to modulate the composition of microbial communities to their advantage in order to adapt to changing biotic and abiotic conditions (Bulgarelli et al. 2012). These strategies involve the ability to favour or select for a beneficial microbiome while avoiding pathogens from the surrounding environment (Bulgarelli et al. 2012; Bahram et al. 2021). There is also evidence that plants and mycorrhizal fungi prefer associating with partners that provide greater benefits for both partners (Bever et al. 2009; Kiers et al. 2011; Bogar et al. 2019), thereby maintaining their interactions under various environmental conditions. Such selective forces by both partners are reflected in the strong association between host identity or genotype and fungal community structure (Gehring et al. 2014; Martínez-García et al. 2015; Küngas, Bahram and Põldmaa 2020; Leopold and Busby 2020). Host specificity of both pathogenic and mutualistic fungi can be reflected in their genomes (Baroncelli et al. 2016; Lofgren et al. 2020). Nevertheless, the strength of these interactions can depend on the presence of a third interacting partner (Lehtonen et al. 2005) as well as environmental conditions and resource availability (Johnson et al. 2015). For example, positive feedback mechanisms in plant–microbial systems may be enhanced by mutualistic interactions of mycorrhizal fungi and N-fixing bacteria (Palakurty, Stinchcombe and Afkhami 2018), which can synergistically increase the fitness of all interacting partners (Afkhami, Friesen and Stinchcombe 2021). Due to the integrated nature of many of these associations between plants and microbes, notably mutualistic fungi, the plant holobiont has been suggested to be a unit of biological organization and selection (Vandenkoornhuyse et al. 2015); however, it is an open question whether such a level of organization can be considered through a macroecological perspective, i.e. can a holobiont disperse as a unit or is it determined by environmental selection following independent dispersal processes of the macro and microorganisms involved? Yet both antagonisms, mutualisms, as well as commensalisms, between land plants and fungi are known to contribute to fungal and plant biogeography (Berbee, James and Strullu-Derrien 2017; Lutzoni et al. 2018; Delavaux et al. 2019), and these interactions strongly impact terrestrial biogeochemical cycles and community dynamics (Tedersoo, Bahram and Zobel 2020).
Fungal-animal interactions
Although the interactions of fungi with animal hosts remain relatively less known, compared to plants, there is also evidence revealing interesting dynamics. For example, animal associated fungal pathogens are one of the most generalist groups of pathogens in terms of host and habitat requirements, highlighted by evidence for the exchange of functional fungi between ecosystems in the case of the transfer of pathogens from human to plant and vice versa (Gauthier and Keller 2013). Such transfers may be facilitated by vectors such as mosquitoes, which are known to carry human opportunistic fungal pathogens existing in non-human environments (Bozic et al. 2017). In addition, some fungal lineages show evolutionary evidence of frequent host switching between plants and insects (Sung, Poinar and Spatafora 2008) and between different insect lineages (e.g. between beetles and ants Araújo and Hughes 2019). There is also evidence that a pathogenic fungus (Erysiphe alphitoides) can interact with various insect herbivores, with a wide range of positive to negative ecological outcomes for the interacting insect depending on species identity, which can affect the structure of insect communities and their interactions with plants at the landscape scale (Tack, Gripenberg and Roslin 2012). A more recent study provides further evidence that pathogenic fungi can mediate the interaction between insect herbivores from competition to facilitation (Klutsch et al. 2016). While the exact mechanisms of the mediating role of fungi remains unknown, besides their direct interactions, fungi may also indirectly interact with insect herbivores by inducing defence signalling pathways and altering the quantity and quality of resources and volatiles in the host plant (Rostás et al. 2006; Klutsch et al. 2016; Bastías et al. 2018; Eberl et al. 2018; Büttner et al. 2021). Also, fungal tissues can provide a richer source of nitrogen and amino acids compared to plant tissues and may thus attract herbivores (Biedermann and Vega 2020; Eberl et al. 2020). Indeed insect–fungus mutualisms—where fungi provide a nutritional source for insects, such as ambrosia beetles, attine ants and termites, and in turn fungi receive protection and dispersal services from insects—is widespread across insect and fungal lineages, with important roles in ecosystem functions (reviewed in Biedermann and Vega 2020). The interaction of plant-insect-fungus also extends to mycorrhizal fungi, where mycorrhizal associations can modify the production and composition of terpenoids, which may contribute significantly to the plant defence response against herbivore attacks (Sharma, Anand and Kapoor 2017). Another example of fungal–insect–host interactions is hyperparasitism, where fungi parasitize parasitic insects, e.g. Laboulbeniales, associated with bat flies (Jensen et al. 2019).
In aquatic ecosystems, perhaps the most well-known groups are notorious aquatic fungal pathogens and parasites, namely chytrids, which are the dominant fungal group in aquatic environments (Bahram et al. 2018) and are the key agent behind the worldwide amphibian decline (Alan Pounds et al. 2006). These fungi can have a broad ecological impact, as they alter aquatic ecosystem stability and functions by acting as a trophic link by driving energy flows to plankton and can regulate their outbreaks (Grami et al. 2011; Rasconi, Niquil and Sime-Ngando 2012; Haraldsson et al. 2018). The zoospores of the parasitic chytrid associated with amphibians, Batrachochytrium dendrobatidis (Bd), are a rich food source for aquatic microfauna and protist grazers, who can control pathogen abundance and infection intensity (Schmeller et al. 2014). There is evidence that Bd can disperse from aquatic to terrestrial systems and retain similar functions (Kolby et al. 2015), and that non-amphibian animals can host and act as a reservoir for Bd (e.g. reptiles (Kilburn, Ibáñez and Green 2011), nematodes (Garmyn et al. 2012) and larval fish (Liew et al. 2017)). Furthermore, the hyperparasitism of parasitic chytrids by other fungi together with fungal-parasitized algae can transfer carbon to the detritus pool, where it can be grazed by protists and metazoans, leading to increased food-web stability in aquatic habitats (reviewed in Gleason et al. 2014; Grossart et al. 2019). Much remains to be discovered about the ecology and biogeography of aquatic fungi given the recent discovery of diverse clades (Jones et al. 2011; Grossart et al. 2019).
Fungal-microbial interactions
In addition to direct interactions with plant and animal hosts, fungi can indirectly interact with their hosts and affect ecosystem functions by altering microbial communities, owing to the often-shared resources and thereby interactions of fungi and bacteria. Such cross-kingdom interactions have been proposed to greatly affect the structure and function of microbiomes with implications for host functioning and health (McFrederick, Mueller and James 2014; Bahram et al. 2018; Deveau et al. 2018; Durán et al. 2018; Bonfante, Venice and Lanfranco 2019; Netherway et al. 2021). For instance, interactions with bacteria may disproportionately affect the growth rate and ecosystem functioning of different fungal guilds (Hiscox et al. 2015; Bahram et al. 2018; Sterkenburg et al. 2018). Competition for nitrogen between EcM fungi and decomposers may lead to greater carbon storage (Averill, Turner and Finzi 2014; Sterkenburg et al. 2018), while stimulation of decomposers via root and mycorrhizal carbon rich exudates may lead to lower carbon storage (Lang et al. 2021). In turn, bacteria can utilize fungal-derived substrates following the decomposition of the more recalcitrant organic matter such as cellulose and lignin (Boer et al. 2005). Based on mapping metabolic interactions using advanced omics methods (e.g. metabolomics; the comprehensive quantitative analysis of all metabolites in a biological system) on the spatiotemporal dynamics of the kefir community, a recent study suggests that metabolic cooperation (i.e. cross-feeding) and spatiotemporal niche partitioning between yeasts and bacteria promote their coexistence in using resources (Blasche et al. 2021). Also, fungal hyphae can act as a dispersal vector, as well as a source of water and nutrients for bacteria (Warmink, Nazir and Elsas 2009; Furuno et al. 2010; Worrich et al. 2017), and thus potentially affect the composition and diversity of bacterial communities. Via transfer of water and nutrients across their mycelial networks, filamentous fungi may be able to better cope with environmental changes than bacteria (Whiteside et al. 2019) and thereby contribute to the stability of bacterial communities. There is also growing evidence that fungi can alter soil bacterial communities through direct competition over resources or production of secondary metabolites (McFrederick, Mueller and James 2014; Bahram et al. 2018; Pierce et al. 2021). In the gut environment—despite acting as a pathogen under certain circumstances—Candida albicans has been shown to act as a mutualistic fungus by inducing antibody production against other fungal infections (Doron et al. 2021).
Fungal-associated bacteria may, in turn, have several beneficial effects on fungi and potentially their host plants or animals. Endosymbiotic bacteria in plant symbiotic fungi (Bonfante and Desiro 2017) could contribute to increased fitness of the fungus (Salvioli et al. 2016). These bacteria play an important role in the co-evolution and functioning of host-fungal systems by facilitating the establishment of symbiotic relationships (e.g. as mycorrhizal helpers) as well as the exchange of genetic material and nutrients between them (reviewed in Bonfante, Venice and Lanfranco 2019). A recent study shows that by using anthelmintic metabolites, endosymbiotic bacteria (Mycoavidus) of a plant growth–promoting fungal species (Mortierella sp.) protect the fungus against nematode attack (Büttner et al. 2021). Insect-fungal mutualistic interactions may also involve bacteria as a third partner. For example, in a beetle-fungal mutualism where a fungus (Entomocorticium sp.) provides food resources for southern pine beetles (Dendroctonus frontalis), bacteria protect the beetle's mutualistic fungus from an antagonistic fungus (Ophiostoma minus) by producing antimicrobials (Scott et al. 2008).
Fungal-viral interactions
Although less characterized, there is also some evidence for fungal–viral interactions in non-fungal hosts. For instance, pulmonary fungal pathogens can impair immune responses to viral infection in humans (Seelbinder et al. 2020), while they can suppress a viral disease (tomato yellow leaf curl virus) in tomato plants (Sun et al. 2021). Other studies also report correlations between the severity or clinical symptoms of viral diseases and fungal diversity, such as in the gut of chronic hepatitis B patients (Chen et al. 2011) as well as the gut and oral samples of COVID-19 patients (Lv et al. 2021; Soffritti et al. 2021). Furthermore, viruses may use fungal mycelial networks to transfer across host systems, as evidenced by the existence of mycoviruses in various tissues of saprotrophic/pathogenic Armillaria species (Linnakoski et al. 2021), between neighbouring EcM fungal clones (Sutela and Vainio 2020), and possibly between different fungal species from different ecological guilds, i.e. saprotrophic, pathogenic and mycorrhizal, within the same forest stand (Vainio et al. 2017). Given the strong interactions between bacteria and fungi in various habitats and through endophytic associations, it is also plausible that fungi interact with bacteriophages. This has at least been shown between the commonly co-occurring human pathogens Pseudomonas aeruginosa (a bacterium) and Aspergillus fumigatus (a fungus), where a bacteriophage that infects P. aeruginosa was found to attach to the surface of and inhibit the metabolic activity of A. fumigatus—through the bacteriophages ability to sequester iron and render it unavailable to the fungus—thus potentially reducing the co-infection of P. aeruginosa and A. fumigatus (Penner et al. 2016). Clearly the interaction of fungi and viruses is an exciting and emerging avenue of research that will provide novel insights into the functioning of biological and ecological systems.
Finally, given that fungi frequently interact with one another, exemplified by the competition and antagonism between different fungal guilds (Rodriguez Estrada et al. 2012; Mukherjee et al. 2014; Fernandez and Kennedy 2016; Thakur et al. 2019) as well as within and between species from the same guild (Yan et al. 2015), there could be multiple fungal species simultaneously involved and interacting in symbiosis or parasitism with a host. Evidence for this comes from the lichen symbiosis where two unrelated fungi ubiquitously associate with algae (Spribille et al. 2016), or from trees capable of hosting both AM and EcM fungal symbionts on the same section of roots (Teste, Jones and Dickie 2020). There is also evidence that pathogenic fungi facilitate each other in infecting hosts, e.g. by providing an adhesion site and promoting co-colonization of vaginal and oral tissues by different Candida species (Alves et al. 2014; Tati et al. 2016). Another example is hyperbiotrophy in fungi, where parasitic fungi of fungi (mycoparasites) boost the virulence of pathogenic fungi to exploit them as a conduit to extract nutrients from their common hosts (Laur et al. 2018).
Taken together, there appears to be a dynamic involvement of multiple species in the pathogenic and symbiotic interactions of fungi with other organisms, with important implications for the health of host animals and plant, as well as ecosystem functioning.
FUNGAL INTERACTIONS IN A CHANGING ENVIRONMENT
Given the great versatility of biotic interactions involving fungi as discussed above, it is unclear exactly how global change factors such as warming and altered precipitation may influence the functioning of these interactions. It is probable that such environmental changes may have important consequences for ecosystem functions such as carbon storage (Clemmensen et al. 2013; Averill and Hawkes 2016) as well as plant responses to biotic stress, which can be contingent on their associated fungi (Bennett and Bever 2007). For example, through mycorrhizal networks, plants can alert their neighbouring plants about herbivory attacks (Babikova et al. 2013), possibly resulting in a less severe response to herbivore attacks with increasing mycorrhizal colonization (Bacht et al. 2019; Sveen et al. 2021). Another example is the tripartite symbiosis between plant-fungus-virus, where plant-fungal symbioses mediated by fungal viruses can confer heat resistance in plants and enable both interacting partners to grow under elevated temperatures (Márquez et al. 2007). Climate warming has been shown to accelerate fungal infection rates, which may alter the dynamics of the food-web structure in aquatic environments (Frenken et al. 2016).
Both climatic and human-induced disturbances may facilitate the emergence of invasive fungi, partly owing to the versatility of fungal interactions and ecology. Fungal invasions can cause major economic losses, with important ecological consequences. This is exemplified by recent shifts in the distributional ranges of several pathogenic fungi towards higher latitudes, posing a threat to production systems in colder climates (Bebber, Ramotowski and Gurr 2013). In addition, given that mycorrhizal fungi probably helped plants to colonize the land, it is not surprising that they still assist, if not play a primary role in facilitating plants in colonizing novel geographic areas (Pringle et al. 2009). For instance, the spread of introduced EcM tree species out of production landscapes and into natural landscapes is associated with both their interaction with co-introduced EcM fungi and through forming novel EcM associations with native EcM fungi (Vlk et al. 2020). Nevertheless, when introduced to new habitats, the lack of co-evolutionary histories may lead to contrasting interaction outcomes for some fungi. A notable example of this is Geomyces destructans that is a widespread commensal fungus in hibernating bats in its native range (Europe), while it causes white-nose syndrome in bats where the fungus is invasive (N. America) (Wibbelt et al. 2010). Furthermore, the versatility of fungal interactions may result in various ecological outcomes from a fungal invasion ranging from increased to reduced fitness, as has been shown in the case of the interaction between the invasive powdery mildew Erysiphe alphitoides with local insect herbivores (Tack, Gripenberg and Roslin 2012). Even when the same symbiosis type persists, the interaction efficiency may be altered when novel interactions are formed with invasive fungi. This is exemplified by the lowered diversity of AM fungi when an exotic AM fungus is introduced to the host plant (Koch et al. 2011). The ecological implications of newly formed interactions by invasive fungi warrants further investigation.
Moving to the health of humans, there is evidence that changes in gut fungal and bacterial communities can affect one another, likely due to their direct interactions. For example, the overgrowth of commensal Candida post-antibiotic treatment (Dollive et al. 2013) can prevent the recovery of bacterial communities resulting in long-term compositional shifts (Mason et al. 2012). Both fungal-bacterial and fungal-fungal interactions in healthy individuals are important to prevent transition of the commensal Candida to a pathogenic state (Limon, Skalski and Underhill 2017). Carbohydrate substrates of fungi play an important role in their fitness in the gut, as evidenced by increased abundance of Candida following the consumption of carbohydrate rich foods, whereas they respond negatively to saturated fatty acids (Hoffmann et al. 2013). In addition, by providing a food source (glycan) and a binding/attachment site, Candida have been suggested to facilitate recolonization of the gut Bacteroidetes during antibiotic recovery (Mason et al. 2012). Finally, given that human pathogens from clinical settings can exist in the environment such as soil (Bensasson et al. 2019; Opulente et al. 2019; O'Brien et al. 2021), it remains to be seen how changes in the environment can affect the evolution of their virulence and resistance traits as well as transmission to humans. By considering a wider range of fungal traits and their role in biotic interactions (Fig. 2), we may be able to better address such questions.
Overall, evidence suggests that while certain fungal interactions may shift in response to environmental changes, leading to altered host and ecosystem functions, some fungi may play a mediating role for their hosts and ecosystems in coping with or succumbing to environmental stress.
CONCLUSIONS AND FUTURE DIRECTIONS
Unique combinations of fungal traits such as diverse dispersal mechanisms as well as versatile genomic, lifestyle and ecophysiological traits could have contributed to their success in spanning multiple scales and dimensions of time, space, and biological interactions, that is not rivalled by other organism groups (Figs. 1, 2). In addition, fungi ubiquitously form complex multikingdom and multispecies interactions, with important implications for ecosystem and host functioning. Several lines of evidence also suggest that fungi act as mediating links between a wide range of organisms and ecosystems, and that both symbioses, parasitism, and indirect interactions have strongly influenced the evolution and distribution patterns of fungi and their interacting taxa (Fig. 3). We conclude that the versatility, complex interactions, and mediating role of fungi further underscores their importance within and across various ecosystems. Yet, the exact underlying mechanisms of the role of fungi in many systems warrant further investigation. In this respect, the key questions that need to be addressed are: How are interactions between fungi with other organisms established? How does evolutionary history influence these interactions and what functional traits facilitate them? How do fungal interactions with other organisms persist over time and space and what conditions facilitate these interactions? What are the relative roles of bacteria and viruses—including HGT and HVT—in fungal interactions with eukaryotic hosts? Further field and controlled experiments are needed to examine to what extent the environment and spatiotemporal variation in fungal communities contributes to the dynamics of fungal interactions. Deciphering links between fungal diversity, fungal and host functional traits and their interactions will not only improve our understanding of the resilience of ecosystems against environmental perturbation, but also help predict future responses to global change including potential range expansions of both fungi and their interacting partners.
Studies of fungal ecology and biogeography, and inferred interactions, have been greatly facilitated by recently developed tools and databases for assigning fungal traits (Nguyen et al. 2016; Põlme et al. 2020; Zanne et al. 2020). Yet, we still need tools and analytical approaches that account for the flexibility in fungal lifestyles and interactions, embrace the complexity of fungi and their influence in ecosystems, and which can reveal realized interactions in mixed fungal communities (Fig. 2). Recent developments in single cell microbial genomics and droplet microfluidics hold great promise for unravelling microbe–microbe and microbe–environment interactions, including communication, competition, cooperation, and food-web dynamics and formation in constructed and controlled micro ecosystems (Nagy et al. 2018). Although these methods are not currently well-suited for studying filamentous fungi and macro eukaryotes due to technical limitations (Millet et al. 2019), there is great potential to adapt these approaches to build custom made systems that mimic small scale heterogeneous soil environments and also microcosms involving seedlings and root symbionts (Aleklett et al. 2018). Recent studies have made exciting progress in this regard, where co-culturing of a plant-associated fungus and bacterium isolated from a host was combined with a novel microfluidics system, comparative metabolomics and analytical methods to identify events of metabolic exchange that initiate a symbiosis between these organisms (Uehling et al. 2019; Büttner et al. 2021).
Genomics data combined with transcriptomics and epigenomics approaches (to study all expressed genes and epigenetic factors in environmental samples, respectively) can provide unique opportunities in the next few years for studying fungal interactions at an unprecedented scale and resolution. Yet, perhaps of key interest to study fungal activity and interactions with hosts, viruses, microbes and ecosystems is metabolomics. In this respect, unravelling metabolic interactions by integrating different omics methods (metagenomics, transcriptomics and metabolomics) and mathematical modelling may be a promising approach to relate species-species interactions, genotypes and phenotypes and to gain mechanistic insights into fungal interactions (Blasche et al. 2021). Increasing genetic, transcriptomic and genomic studies on molecular host-fungus interactions and biomolecule degradation pathways provide comprehensive characterisation of functional gene families and activity centres of enzymes, transporters, signalling molecules and transposable elements.
By moving towards the study of the phenotypic expression of fungi, host systems and their interactions, and ecosystems, can we begin to collect cause and effect information on fungal interactions and functional outcomes. By exploring fungi beyond traditional disciplinary and ecosystem boundaries, can we further unravel how communities of fungi and their interacting organisms may shift in response to changes in environmental conditions and how this may affect host and ecosystem functioning and health.
ACKNOWLEDGEMENTS
The authors apologize to colleagues whose work was not discussed in this review. The authors thank Jan Bengtsson and three anonymous reviewers for constructive comments on an earlier version of the manuscript.
Contributor Information
Mohammad Bahram, Department of Ecology, Swedish University of Agricultural Sciences, Uppsala, Ulls väg 16, 756 51 Sweden; Institute of Ecology and Earth Sciences, University of Tartu, 40 Lai St., 51005, Tartu, Estonia.
Tarquin Netherway, Department of Ecology, Swedish University of Agricultural Sciences, Uppsala, Ulls väg 16, 756 51 Sweden.
FUNDING
This work was supported by the Swedish Research Councils Vetenskapsrådet (Grants 2017–05019 and 2021–03724) and Formas (Grant 2020–00807).
Conflicts of interest
None declared.
REFERENCES
- Aanen DK, Hoekstra RF. Why sex is good: on fungi and beyond. Sex in Fungi: molecular determination and evolutionary implications. Washington DC, USA: ASM press, 2007, 527–34. [Google Scholar]
- Afkhami ME, Friesen ML, Stinchcombe JR. Multiple mutualism effects generate synergistic selection and strengthen fitness alignment in the interaction between legumes, rhizobia and mycorrhizal fungi. Ecology Letters. 2021;24:1824–34. [DOI] [PubMed] [Google Scholar]
- Alan Pounds J, Bustamante MR, Coloma LAet al. . Widespread amphibian extinctions from epidemic disease driven by global warming. Nature. 2006;439:161–7. [DOI] [PubMed] [Google Scholar]
- Aleklett K, Kiers ET, Ohlsson Pet al. . Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 2018;12:312–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CT, Wei X-Q, Silva Set al. . Candida albicans promotes invasion and colonisation of candida glabrata in a reconstituted human vaginal epithelium. J Infect. 2014;69:396–407. [DOI] [PubMed] [Google Scholar]
- Anderson JB, Bruhn JN, Kasimer Det al. . Clonal evolution and genome stability in a 2500-year-old fungal individual. Procee Royal Soc B: Biolog Sci. 2018;285:20182233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika IB, Wei S, Cao Cet al. . Phytopathogenic fungus hosts a plant virus: a naturally occurring cross-kingdom viral infection. Proc Natl Acad Sci. 2017;114:12267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo JPM, Hughes DP. Zombie-Ant fungi emerged from Non-manipulating, beetle-infecting ancestors. Curr Biol. 2019;29:3735–8. [DOI] [PubMed] [Google Scholar]
- Archer S, Lee K, Caruso T. et al. Diverse Recruitment to a Globally Structured Atmospheric Microbiome. In Review, 2021. [Google Scholar]
- Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots?. Ecology. 2007;88:541–9. [DOI] [PubMed] [Google Scholar]
- Averill C, Hawkes CV. Ectomycorrhizal fungi slow soil carbon cycling. Bardgett R (ed.). Ecology Letters. 2016;19:937–47. [DOI] [PubMed] [Google Scholar]
- Averill C, Turner BL, Finzi AC. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature. 2014;505:543. [DOI] [PubMed] [Google Scholar]
- Babikova Z, Gilbert L, Bruce TJet al. . Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecology Letters. 2013;16:835–43. [DOI] [PubMed] [Google Scholar]
- Bacht M, Tarkka MT, López IFet al. . Tree response to herbivory is affected by endogenous rhythmic growth and attenuated by cotreatment with a mycorrhizal fungus. Mol Plant-Microbe Inter®. 2019;32:770–81. [DOI] [PubMed] [Google Scholar]
- Bagchi R, Gallery RE, Gripenberg Set al. . Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature. 2014;506:85–8. [DOI] [PubMed] [Google Scholar]
- Bahram M, Hildebrand F, Forslund SKet al. . Structure and function of the global topsoil microbiome. Nature. 2018;560:233–7. [DOI] [PubMed] [Google Scholar]
- Bahram M, Netherway T, Frioux Cet al. . Metagenomic assessment of the global diversity and distribution of bacteria and fungi. Environ Microbiol. 2021;23:316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroncelli R, Amby DB, Zapparata Aet al. . Gene family expansions and contractions are associated with host range in plant pathogens of the genus colletotrichum. BMC Genomics. 2016;17:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastías DA, Martínez-Ghersa MA, Newman JAet al. . Jasmonic acid regulation of the anti-herbivory mechanism conferred by fungal endophytes in grasses. J Ecol. 2018;106:2365–79. [Google Scholar]
- Bebber DP, Ramotowski MA, Gurr SJ. Crop pests and pathogens move polewards in a warming world. Nature Climate Change. 2013;3:985. [Google Scholar]
- Becker J, Liermann JC, Opatz Tet al. . GKK1032A 2, a secondary metabolite from penicillium sp. IBWF-029-96, inhibits conidial germination in the rice blast fungus magnaporthe oryzae. J Antibiot (Tokyo). 2012;65:99–102. [DOI] [PubMed] [Google Scholar]
- Bennett AE, Bever JD. Mycorrhizal species differentially alter plant growth and response to herbivory. Ecology. 2007;88:210–8. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Dicks J, Ludwig JMet al. . Diverse lineages of candida albicans live on old oaks. Genetics. 2019;211:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, James TY, Strullu-Derrien C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Annu Rev Microbiol. 2017;71:41–60. [DOI] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BMet al. . Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett. 2009;12:13–21. [DOI] [PubMed] [Google Scholar]
- Bian R, Andika IB, Pang Tet al. . Facilitative and synergistic interactions between fungal and plant viruses. Proc Natl Acad Sci. 2020;117:3779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann PH, Vega FE. Ecology and evolution of insect–fungus mutualisms. Annu Rev Entomol. 2020;65:431–55. [DOI] [PubMed] [Google Scholar]
- Blasche S, Kim Y, Mars RATet al. . Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nature Microbiol. 2021;6:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer W de, Folman LB, Summerbell RCet al. . Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. [DOI] [PubMed] [Google Scholar]
- Bogar L, Peay K, Kornfeld Aet al. . Plant-mediated partner discrimination in ectomycorrhizal mutualisms. Mycorrhiza. 2019;29:97–111. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Desirò A. Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. ISME J. 2017;11:1727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Venice F, Lanfranco L. The mycobiota: fungi take their place between plants and bacteria. Curr Opin Microbiol. 2019;49:18–25. [DOI] [PubMed] [Google Scholar]
- Bonneville S, Delpomdor F, Préat Aet al. . Molecular identification of fungi microfossils in a neoproterozoic shale rock. Sci Adv. 2020;6:eaax7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouws H, Wattenberg A, Zorn H. Fungal secretomes—nature's toolbox for white biotechnology. Appl Microbiol Biotechnol. 2008;80:381. [DOI] [PubMed] [Google Scholar]
- Bozic J, Capone A, Pediconi Det al. . Mosquitoes can harbour yeasts of clinical significance and contribute to their environmental dissemination. Environ Microbiol Rep. 2017;9:642–8. [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018;220:1108–15. [DOI] [PubMed] [Google Scholar]
- Bruto M, Prigent-Combaret C, Luis Pet al. . Frequent, independent transfers of a catabolic gene from bacteria to contrasted filamentous eukaryotes. Proc R Soc B Biol Sci. 2014;281:20140848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi Ket al. . Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–5. [DOI] [PubMed] [Google Scholar]
- Büttner H, Niehs SP, Vandelannoote Ket al. . Bacterial endosymbionts protect beneficial soil fungus from nematode attack. Proc Natl Acad Sci. 2021;118:e2110669118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiafa MV, Jusino MA, Wilkie ACet al. . Discovering the role of patagonian birds in the dispersal of truffles and other mycorrhizal fungi. Curr Biol. 10.2139/ssrn.3854514. [DOI] [PubMed] [Google Scholar]
- Calo S, Billmyre RB, Heitman J. Generators of phenotypic diversity in the evolution of pathogenic microorganisms. In: True-Krob H (ed). PLoS Pathog. 2013;9:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EK, Martins IS, Lavelle Pet al. . Global mismatches in aboveground and belowground biodiversity. Conserv Biol. 2019;33:1187–92. [DOI] [PubMed] [Google Scholar]
- Castanera R, López-Varas L, Borgognone Aet al. . Transposable elements versus the fungal genome: impact on whole-genome architecture and transcriptional profiles. PLos Genet. 2016;12:e1006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda A, Roze LV, Kang Set al. . A key role for vesicles in fungal secondary metabolism. Proc Natl Acad Sci. 2009;106:19533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Swenson NG, Ji Net al. . Differential soil fungus accumulation and density dependence of trees in a subtropical forest. Science. 2019;366:124–8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen Z, Guo Ret al. . Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis b virus infection. Diagn Microbiol Infect Dis. 2011;70:492–8. [DOI] [PubMed] [Google Scholar]
- Choi J, Park J, Kim Det al. . Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics. 2010;11:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge AW. Ecological role of hypogeous ectomycorrhizal fungi in australian forests and woodlands. In: Diversity and Integration in Mycorrhizas, Dordrecht: Springer, 2002, 291–305. [Google Scholar]
- Clemmensen KE, Bahr A, Ovaskainen Oet al. . Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science. 2013;339:1615–8. [DOI] [PubMed] [Google Scholar]
- Coleine C, Stajich JE, Ríos A de loset al. . Beyond the extremes: rocks as ultimate refuge for fungi in drylands. Mycologia. 2021;113:108–33. [DOI] [PubMed] [Google Scholar]
- Cordero RJ, Casadevall A. Functions of fungal melanin beyond virulence. Fungal Biol Rev. 2017;31:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J, Moora M, Opik Met al. . Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science. 2015;349:970–3. [DOI] [PubMed] [Google Scholar]
- Delavaux CS, Weigelt P, Dawson Wet al. . Mycorrhizal fungi influence global plant biogeography. Nat Ecol Evol. 2019;3:424–9. [DOI] [PubMed] [Google Scholar]
- Deveau A, Bonito G, Uehling Jet al. . Bacterial-Fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. [DOI] [PubMed] [Google Scholar]
- Dollive S, Chen Y-Y, Grunberg Set al. . Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One. 2013;8:e71806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron I, Leonardi I, Li XVet al. . Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell. 2021;184:1017–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán P, Thiergart T, Garrido-Oter Ret al. . Microbial interkingdom interactions in roots promote arabidopsis survival. Cell. 2018;175:973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer O. Covid-19: india sees record deaths as “black fungus” spreads fear. BMJ. 2021;373:n1238. [DOI] [PubMed] [Google Scholar]
- Dyer PS, Kück U. Sex and the imperfect fungi. Microbiol Spectr. 2017;5:5–3. [DOI] [PubMed] [Google Scholar]
- Eberl F, Bobadilla MF de, Reichelt Met al. . Herbivory meets fungivory: insect herbivores feed on plant pathogenic fungi for their own benefit. Ecol Lett. 2020;23:1073–84. [DOI] [PubMed] [Google Scholar]
- Eberl F, Hammerbacher A, Gershenzon Jet al. . Leaf rust infection reduces herbivore-induced volatile emission in black poplar and attracts a generalist herbivore. New Phytol. 2018;220:760–72. [DOI] [PubMed] [Google Scholar]
- Ehinger MO, Croll D, Koch AMet al. . Significant genetic and phenotypic changes arising from clonal growth of a single spore of an arbuscular mycorrhizal fungus over multiple generations. New Phytol. 2012;196:853–61. [DOI] [PubMed] [Google Scholar]
- Faino L, Seidl MF, Shi-Kunne Xet al. . Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 2016;26:1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CW, Kennedy PG. Revisiting the ‘Gadgil effect’: do interguild fungal interactions control carbon cycling in forest soils?. New Phytol. 2016;209:1382–94. [DOI] [PubMed] [Google Scholar]
- Francis AP, Currie DJ. A globally consistent richness-climate relationship for angiosperms. Am Nat. 2003;161:523–36. [DOI] [PubMed] [Google Scholar]
- Frantzeskakis L, Kracher B, Kusch Set al. . Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics. 2018;19:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenken T, Velthuis M, Domis LN de Set al. . Warming accelerates termination of a phytoplankton spring bloom by fungal parasites. Global Change Biol. 2016;22:299–309. [DOI] [PubMed] [Google Scholar]
- Fricker MD, Heaton LLM, Jones NSet al. . The mycelium as a network. Microbiol Spectr . 2017;5. DOI: 10.1128/microbiolspec.FUNK-0033-2017. [DOI] [PubMed] [Google Scholar]
- Furuno S, Päzolt K, Rabe Cet al. . Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ Microbiol. 2010;12:1391–8. [DOI] [PubMed] [Google Scholar]
- Gan T, Luo T, Pang Ket al. . Cryptic terrestrial fungus-like fossils of the early ediacaran period. Nat Commun. 2021;12:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmyn A, Van Rooij P, Pasmans Fet al. . Waterfowl: potential environmental reservoirs of the chytrid fungus batrachochytrium dendrobatidis. Fisher m (Mat) c (ed).PLoS One. 2012;7:e35038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier GM, Keller NP. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol. 2013;61:146–57. [DOI] [PubMed] [Google Scholar]
- Gehring C, Flores‐Rentería D, Sthultz CMet al. . Plant genetics and interspecific competitive interactions determine ectomycorrhizal fungal community responses to climate change. Mol Ecol. 2014;23:1379–91. [DOI] [PubMed] [Google Scholar]
- Genre A, Lanfranco L, Perotto Set al. . Unique and common traits in mycorrhizal symbioses. Nat Rev Microbiol. 2020;18:649–60. [DOI] [PubMed] [Google Scholar]
- Girard V, Dieryckx C, Job Cet al. . Secretomes: the fungal strike force. Proteomics. 2013;13:597–608. [DOI] [PubMed] [Google Scholar]
- Gleason FH, Lilje O, Marano AVet al. . Ecological functions of zoosporic hyperparasites. Frontiers in Microbiology. 2014;5:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan JJ, Pringle A. Long-Distance dispersal of fungi. Microbiology Spectrum. 2017;5:5.4.03. [DOI] [PubMed] [Google Scholar]
- Grami B, Rasconi S, Niquil Net al. . Functional effects of parasites on food web properties during the spring diatom bloom in lake pavin: a linear inverse modeling analysis. PLoS One. 2011;6:e23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossart H-P, Van den Wyngaert S, Kagami Met al. . Fungi in aquatic ecosystems. Nat Rev Microbiol. 2019;17:339–54. [DOI] [PubMed] [Google Scholar]
- Gusareva ES, Acerbi E, Lau KJXet al. . Microbial communities in the tropical air ecosystem follow a precise diel cycle. Proc Natl Acad Sci. 2019;116:23299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsson M, Gerphagnon M, Bazin Pet al. . Microbial parasites make cyanobacteria blooms less of a trophic dead end than commonly assumed. The ISME Journal. 2018;12:1008–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiology Spectrum. 2017;5:5.4.10. [DOI] [PubMed] [Google Scholar]
- Hess J, Skrede I, Chaib De Mares Met al. . Rapid divergence of genome architectures following the origin of an ectomycorrhizal symbiosis in the genus amanita. Mol Biol Evol. 2018;35:2786–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Skrede I, Wolfe BEet al. . Transposable element dynamics among asymbiotic and ectomycorrhizal amanita fungi. Genome Biol Evol. 2014;6:1564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillebrand H. On the generality of the latitudinal diversity gradient. Am Nat. 2004;163:192–211. [DOI] [PubMed] [Google Scholar]
- Hiscox J, Savoury M, Vaughan IPet al. . Antagonistic fungal interactions influence carbon dioxide evolution from decomposing wood. Fungal Ecol. 2015;14:24–32. [Google Scholar]
- Hoffmann C, Dollive S, Grunberg Set al. . Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One. 2013;8:e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hull CM. Sporulation: how to survive on planet earth (and beyond). Curr Genet. 2017;63:831–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DP, Wappler T, Labandeira CC. Ancient death-grip leaf scars reveal ant–fungal parasitism. Biol Lett. 2011;7:67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey V, Zscheischler J, Ciais Pet al. . Sensitivity of atmospheric CO 2 growth rate to observed changes in terrestrial water storage. Nature. 2018;560:628–31. [DOI] [PubMed] [Google Scholar]
- Jacobson ES. Pathogenic roles for fungal melanins. Clin Microbiol Rev. 2000;13:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusz G, Pawlik A, Sulej Jet al. . Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev. 2017;41:941–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KM, Rodrigues L, Pape Tet al. . Hyperparasitism in caves: bats, bat flies and ectoparasitic fungus interaction. J Invertebr Pathol. 2019;166:107206. [DOI] [PubMed] [Google Scholar]
- Johnson NC, Wilson GW, Wilson JAet al. . Mycorrhizal phenotypes and the l aw of the m inimum. New Phytol. 2015;205:1473–84. [DOI] [PubMed] [Google Scholar]
- Jones MDM, Forn I, Gadelha Cet al. . Discovery of novel intermediate forms redefines the fungal tree of life. Nature. 2011;474:200–3. [DOI] [PubMed] [Google Scholar]
- Keller NP. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 2019;17:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PG, Matheny PB, Ryberg KMet al. . Scaling up: examining the macroecology of ectomycorrhizal fungi. Mol Ecol. 2012;21:4151–4. [DOI] [PubMed] [Google Scholar]
- Khaldi N, Collemare J, Lebrun M-Het al. . Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 2008;9:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid S, Baccile JA, Spraker JEet al. . NRPS-Derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem Biol. 2018;13:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Yet al. . Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–2. [DOI] [PubMed] [Google Scholar]
- Kilburn VL, Ibáñez R, Green DM. Reptiles as potential vectors and hosts of the amphibian pathogen batrachochytrium dendrobatidis in panama. Dis Aquat Organ. 2011;97:127–34. [DOI] [PubMed] [Google Scholar]
- Kiss E, Hegedüs B, Virágh Met al. . Comparative genomics reveals the origin of fungal hyphae and multicellularity. Nat Commun. 2019;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivlin SN, Lynn JS, Kazenel MRet al. . Biogeography of plant-associated fungal symbionts in mountain ecosystems: a meta-analysis. Diversity Distributions. 2017;23:1067–77. [Google Scholar]
- Klutsch JG, Najar A, Cale JAet al. . Direction of interaction between mountain pine beetle (Dendroctonus ponderosae) and resource-sharing wood-boring beetles depends on plant parasite infection. Oecologia. 2016;182:1–12. [DOI] [PubMed] [Google Scholar]
- Kobmoo N, Mongkolsamrit S, Tasanathai Ket al. . Molecular phylogenies reveal host-specific divergence of ophiocordyceps unilateralis sensu lato following its host ants. Mol Ecol. 2012;21:3022–31. [DOI] [PubMed] [Google Scholar]
- Koch AM, Antunes PM, Kathryn Barto Eet al. . The effects of arbuscular mycorrhizal (AM) fungal and garlic mustard introductions on native AM fungal diversity. Biological Invasions. 2011;13:1627–39. [Google Scholar]
- Kolby JE, Ramirez SD, Berger Let al. . Terrestrial dispersal and potential environmental transmission of the amphibian chytrid fungus (Batrachochytrium dendrobatidis). PLoS One. 2015;10:e0125386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov Ket al. . Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. [DOI] [PubMed] [Google Scholar]
- Kreft H, Jetz W. Global patterns and determinants of vascular plant diversity. Proc Natl Acad Sci. 2007;104:5925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küngas K, Bahram M, Põldmaa K. Host tree organ is the primary driver of endophytic fungal community structure in a hemiboreal forest. FEMS Microbiol Ecol. 2020;96:fiz199. [DOI] [PubMed] [Google Scholar]
- Lang AK, Jevon FV, Vietorisz CRet al. . Fine roots and mycorrhizal fungi accelerate leaf litter decomposition in a northern hardwood forest regardless of dominant tree mycorrhizal associations. New Phytol. 2021;230:316–26. [DOI] [PubMed] [Google Scholar]
- Lange L, Barrett K, Meyer AS. New method for identifying fungal kingdom enzyme hotspots from genome sequences. J Fungi. 2021;7:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laur J, Ramakrishnan GB, Labbé Cet al. . Effectors involved in fungal–fungal interaction lead to a rare phenomenon of hyperbiotrophy in the tritrophic system biocontrol agent–powdery mildew–plant. New Phytol. 2018;217:713–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen P, Helander M, Wink Met al. . Transfer of endophyte-origin defensive alkaloids from a grass to a hemiparasitic plant. Ecology Letters. 2005;8:1256–63. [Google Scholar]
- Leopold DR, Busby PE. Host genotype and colonist arrival order jointly govern plant microbiome composition and function. Curr Biol. 2020:S0960982220308277. [DOI] [PubMed] [Google Scholar]
- Li M, Zhao J, Tang Net al. . Horizontal gene transfer from bacteria and plants to the arbuscular mycorrhizal fungus rhizophagusirregularis. Front Plant Sci. 2018;9. DOI: 10.3389/fpls.2018.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nielsen K. Morphology changes in human fungal pathogens upon interaction with the host. J Fungi. 2017;3:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew N, Mazon Moya MJ, Wierzbicki CJet al. . Chytrid fungus infection in zebrafish demonstrates that the pathogen can parasitize non-amphibian vertebrate hosts. Nat Commun. 2017;8:15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Skalski JH, Underhill DM. Commensal fungi in health and disease. Cell Host & Microbe. 2017;22:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnakoski R, Sutela S, Coetzee MPAet al. . Armillaria root rot fungi host single-stranded RNA viruses. Sci Rep. 2021;11:7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Jiang Det al. . Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol. 2010;84:11876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xie J, Cheng Jet al. . Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc Natl Acad Sci. 2016;113:12803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren LA, Nguyen NH, Vilgalys Ret al. . Comparative genomics reveals dynamic genome evolution in host specialist ectomycorrhizal fungi. New Phytol. 2020;230:774–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney BP, Ryberg M, Hampe Fet al. . Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol Ecol. 2016;25:630–47. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Nowak MD, Alfaro MEet al. . Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun. 2018;9:5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Gu S, Jiang Het al. . Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Communications Biology. 2021;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD. Unbelievable but true: epigenetics and chromatin in fungi. Trends Genet. 2021;37:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares MCD, Hess J, Floudas Det al. . Horizontal transfer of carbohydrate metabolism genes into ectomycorrhizal amanita. New Phytol. 2015;205:1552–64. [DOI] [PubMed] [Google Scholar]
- Márquez LM, Redman RS, Rodriguez RJet al. . A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315:513–5. [DOI] [PubMed] [Google Scholar]
- Martín JF. Transport systems, intracellular traffic of intermediates and secretion of β-lactam antibiotics in fungi. Fungal Biol Biotechnol. 2020;7:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García LB, Richardson SJ, Tylianakis JMet al. . Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytol. 2015;205:1565–76. [DOI] [PubMed] [Google Scholar]
- Mason KL, Erb Downward JR, Mason KDet al. . Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80:3371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MK, Whigham DF, Canchani-Viruet A. Mycorrhizal fungi affect orchid distribution and population dynamics. New Phytol. 2018;219:1207–15. [DOI] [PubMed] [Google Scholar]
- McFrederick QS, Mueller UG, James RR. Interactions between fungi and bacteria influence microbial community structure in the megachile rotundata larval gut. Procee Royal Soc B: Biolog Sci. 2014;281:20132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet LJ, Aufrecht J, Labbé Jet al. . Increasing access to microfluidics for studying fungi and other branched biological structures. Fungal Biol Biotechnol. 2019;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter NJ, Buatois LA, Mángano MGet al. . Early bursts of diversification defined the faunal colonization of land. Nat Ecol Evol. 2017;1:0175. [Google Scholar]
- Miyauchi S, Kiss E, Kuo Aet al. . Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun. 2020;11:5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller M, Stukenbrock EH. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol. 2017;15:756–71. [DOI] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J, Retuerto Met al. . Oral mycobiome analysis of HIV-Infected patients: identification of pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014;10:e1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy K, Ábrahám Á, Keymer JEet al. . Application of microfluidics in experimental ecology: the importance of being spatial. Front Microbiol. 2018;9:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo-Ortiz MA, Gabaldón T. Fungal evolution: major ecological adaptations and evolutionary transitions. Biological Rev. 2019;94:1443–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Schüßler A, Bonfante P. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the mollicutes. ISME J. 2010;4:862–71. [DOI] [PubMed] [Google Scholar]
- Nerva L, Varese GC, Falk BWet al. . Mycoviruses of an endophytic fungus can replicate in plant cells: evolutionary implications. Sci Rep. 2017;7:1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherway T, Bengtsson J, Krab EJet al. . Biotic interactions with mycorrhizal systems as extended nutrient acquisition strategies shaping forest soil communities and functions. Basic Appl Ecol. 2021;50:25–42. [Google Scholar]
- Nguyen NH, Song Z, Bates STet al. . FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology. 2016;20:241–8. [Google Scholar]
- Nieuwenhuis BPS, James TY. The frequency of sex in fungi. Philos Transac Royal Soc B: Biolog Sci. 2016;371:20150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Anslan S, Bahram Met al. . Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat Rev Microbiol. 2019;17:95–109. [DOI] [PubMed] [Google Scholar]
- Nuske SJ, Anslan S, Tedersoo Let al. . Ectomycorrhizal fungal communities are dominated by mammalian dispersed truffle-like taxa in north-east australian woodlands. Mycorrhiza. 2019;29:181–93. [DOI] [PubMed] [Google Scholar]
- O'Brien CE, Oliveira-Pacheco J, Cinnéide EÓet al. . Population genomics of the pathogenic yeast candida tropicalis identifies hybrid isolates in environmental samples. PLoS Pathog. 2021;17:e1009138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn NT. Genome Size and Host Specialization in Parasites. Graduate Theses and Dissertations. FL, USA: University of South Florida, 2019. [Google Scholar]
- Opulente DA, Langdon QK, Buh KVet al. . Pathogenic budding yeasts isolated outside of clinical settings. FEMS Yeast Res. 2019;19. DOI: 10.1093/femsyr/foz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek LS van, Saikkonen K. Impact of bacterial–fungal interactions on the colonization of the endosphere. Trends Plant Sci. 2016;21:230–42. [DOI] [PubMed] [Google Scholar]
- Palakurty SX, Stinchcombe JR, Afkhami ME. Cooperation and coexpression: how coexpression networks shift in response to multiple mutualists. Mol Ecol. 2018;27:1860–73. [DOI] [PubMed] [Google Scholar]
- Peay KG, Kennedy PG, Talbot JM. Dimensions of biodiversity in the earth mycobiome. Nat Rev Microbiol. 2016;14:434. [DOI] [PubMed] [Google Scholar]
- Penner JC, Ferreira JAG, Secor PRet al. . Pf4 bacteriophage produced by pseudomonas aeruginosa inhibits aspergillus fumigatus metabolism via iron sequestration. Microbiology. 2016;162:1583–94. [DOI] [PubMed] [Google Scholar]
- Pfannenstiel BT, Keller NP. On top of biosynthetic gene clusters: how epigenetic machinery influences secondary metabolism in fungi. Biotechnol Adv. 2019;37:107345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce EC, Morin M, Little JCet al. . Bacterial–fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nature Microbiology. 2021;6:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozynski KA, Malloch DW. The origin of land plants: a matter of mycotrophism. Biosystems. 1975;6:153–64. [DOI] [PubMed] [Google Scholar]
- Plett JM, Martin F. Reconsidering mutualistic plant–fungal interactions through the lens of effector biology. Curr Opin Plant Biol. 2015;26:45–50. [DOI] [PubMed] [Google Scholar]
- Põlme S, Abarenkov K, Henrik Nilsson Ret al. . FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Diversity. 2020;105:1–16. [Google Scholar]
- Põlme S, Bahram M, Jacquemyn Het al. . Host preference and network properties in biotrophic plant–fungal associations. New Phytol. 2018;217:1230–9. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G. Genetic systems based on hetero caryosis. In: Cold Spring Harbor Symposia on Quantitative Biology, Vol. 11, NY, USA: Cold Spring Harbor Laboratory Press, 1946, 193–201. [Google Scholar]
- Pringle A, Bever JD, Gardes Met al. . Mycorrhizal symbioses and plant invasions. Annu Rev Ecol Evol Syst. 2009;40:699–715. [Google Scholar]
- Rasconi S, Niquil N, Sime-Ngando T. Phytoplankton chytridiomycosis: community structure and infectivity of fungal parasites in aquatic ecosystems. Environ Microbiol. 2012;14:2151–70. [DOI] [PubMed] [Google Scholar]
- Redecker D. Glomalean fungi from the ordovician. Science. 2000;289:1920–1. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass Het al. . Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci. 1994;91:11841–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich MK, Vigneron N, Libourel Cet al. . Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science. 2021;372:864–8. [DOI] [PubMed] [Google Scholar]
- Rodriguez Estrada AE, Jonkers W, Corby Kistler Het al. . Interactions between fusarium verticillioides, ustilago maydis, and zea mays: an endophyte, a pathogen, and their shared plant host. Fungal Genet Biol. 2012;49:578–87. [DOI] [PubMed] [Google Scholar]
- Rohlfs M, Churchill ACL. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet Biol. 2011;48:23–34. [DOI] [PubMed] [Google Scholar]
- Roossinck MJ. Evolutionary and ecological links between plant and fungal viruses. New Phytol. 2019;221:86–92. [DOI] [PubMed] [Google Scholar]
- Rostás M, Ton J, Mauch-Mani Bet al. . Fungal infection reduces herbivore-induced plant volatiles of maize but does not affect naïve parasitoids. J Chem Ecol. 2006;32:1897–909. [DOI] [PubMed] [Google Scholar]
- Salvioli A, Ghignone S, Novero Met al. . Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J. 2016;10:130–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ramírez S, Tulloss RE, Amalfi Met al. . Palaeotropical origins, boreotropical distribution and increased rates of diversification in a clade of edible ectomycorrhizal mushrooms (Amanita section caesareae). J Biogeogr. 2015;42:351–63. [Google Scholar]
- Schmeller DS, Blooi M, Martel Aet al. . Microscopic aquatic predators strongly affect infection dynamics of a globally emerged pathogen. Curr Biol. 2014;24:176–80. [DOI] [PubMed] [Google Scholar]
- Scott JJ, Oh DC, Yuceer MCet al. . Bacterial protection of beetle-fungus mutualism. Science. 2008;322:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelbinder B, Wallstabe J, Marischen Let al. . Triple RNA-Seq reveals synergy in a human virus-fungus Co-infection model. Cell Rep. 2020;33:108389. [DOI] [PubMed] [Google Scholar]
- Sharma E, Anand G, Kapoor R. Terpenoids in plant and arbuscular mycorrhiza-reinforced defence against herbivorous insects. Ann Bot. 2017;119:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh H, Boaz BE, Millar CIet al. . Symbiotic interactions above treeline of long-lived pines: mycorrhizal advantage of limber pine (Pinus flexilis) over great basin bristlecone pine (Pinus longaeva) at the seedling stage. Journal of Ecology. 2020;108:908–16. [Google Scholar]
- Sinn BT, Barrett CF. Ancient mitochondrial gene transfer between fungi and the orchids. Mol Biol Evol. 2020;37:44–57. [DOI] [PubMed] [Google Scholar]
- Smith GR, Finlay RD, Stenlid Jet al. . Growing evidence for facultative biotrophy in saprotrophic fungi: data from microcosm tests with 201 species of wood-decay basidiomycetes. New Phytol. 2017;215:747–55. [DOI] [PubMed] [Google Scholar]
- Soffritti I, D'Accolti M, Fabbri Cet al. . Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: a cross-sectional study. Frontiers in Microbiology. 2021;12:687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spribille T, Tuovinen V, Resl Pet al. . Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science. 2016;353:488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J, Gallone B, Verstrepen KJ. Interspecific hybridization as a driver of fungal evolution and adaptation. Nat Rev Microbiol. 2021;19:485–500. [DOI] [PubMed] [Google Scholar]
- Steidinger BS, Crowther TW, Liang Jet al. . Climatic controls of decomposition drive the global biogeography of forest-tree symbioses. Nature. 2019;569:404–8. [DOI] [PubMed] [Google Scholar]
- Sterkenburg E, Clemmensen KE, Ekblad Aet al. . Contrasting effects of ectomycorrhizal fungi on early and late stage decomposition in a boreal forest. ISME J. 2018;12:2187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strullu-Derrien C, Spencer ART, Goral Tet al. . New insights into the evolutionary history of fungi from a 407 ma blastocladiomycota fossil showing a complex hyphal thallus. Philos Transac Royal Soc B: Biolog Sci. 2018;373:20160502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wang Z, Zhang Zet al. . The roles of entomopathogenic fungal infection of viruliferous whiteflies in controlling tomato yellow leaf curl virus. Biol Control. 2021;156:104552. [Google Scholar]
- Sung G-H, Poinar GO, Spatafora JW. The oldest fossil evidence of animal parasitism by fungi supports a cretaceous diversification of fungal–arthropod symbioses. Mol Phylogenet Evol. 2008;49:495–502. [DOI] [PubMed] [Google Scholar]
- Sutela S, Forgia M, Vainio EJet al. . The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evolution. 2020;6. DOI: 10.1093/ve/veaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutela S, Vainio EJ. Virus population structure in the ectomycorrhizal fungi lactarius rufus and l. tabidus at two forest sites in southern finland. Virus Res. 2020;285:197993. [DOI] [PubMed] [Google Scholar]
- Sveen TR, Netherway T, Juhanson Jet al. . Plant-microbe interactions in response to grassland herbivory and nitrogen eutrophication. Soil Biol Biochem. 2021;156:108208. [Google Scholar]
- Tack AJM, Gripenberg S, Roslin T. Cross-kingdom interactions matter: fungal-mediated interactions structure an insect community on oak. Ecol Lett. 2012;15:177–85. [DOI] [PubMed] [Google Scholar]
- Tati S, Davidow P, McCall Aet al. . Candida glabrata binding to candida albicans hyphae enables its development in oropharyngeal candidiasis. Noverr MC (ed).PLoS Pathog. 2016;12:e1005522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Branco S, Gao Cet al. . Sources of fungal genetic variation and associating it with phenotypic diversity. Microbiol Spec. 2017;5:5.5.06. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Polme Set al. . Global diversity and geography of soil fungi. Science. 2014;346:1256688. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Toots Met al. . Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol Ecol. 2012;21:4160–70. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Zobel M. How mycorrhizal associations drive plant population and community biology. Science. 2020;367. DOI: 10.1126/science.aba1223. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Sánchez-Ramírez S, Koljalg Uet al. . High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Diversity. 2018;90:135–59. [Google Scholar]
- Teste FP, Jones MD, Dickie IA. Dual-mycorrhizal plants: their ecology and relevance. New Phytol. 2020;225:1835–51. [DOI] [PubMed] [Google Scholar]
- Thakur MP, Quast V, Dam NMd vanet al. . Interactions between functionally diverse fungal mutualists inconsistently affect plant performance and competition. Oikos. 2019;128:1136–46. [Google Scholar]
- Tisthammer KH, Cobian GM, Amend AS. Global biogeography of marine fungi is shaped by the environment. Fungal Ecology. 2016;19:39–46. [Google Scholar]
- Torres-Cortés G, Ghignone S, Bonfante Pet al. . Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci. 2015;112:7785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Garcia S, Yaseen I, Shukla Met al. . Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature. 2020;585:453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehling JK, Entler MR, Meredith HRet al. . Microfluidics and metabolomics reveal symbiotic bacterial-fungal interactions between mortierella elongata and burkholderia include metabolite exchange. Front Microbiol. 2019;10:2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S, Xu X, Lowry Det al. . Subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin. Cell Rep. 2016;14:2511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio EJ, Pennanen T, Rajala Tet al. . Occurrence of similar mycoviruses in pathogenic, saprotrophic and mycorrhizal fungi inhabiting the same forest stand. FEMS Microbiol Ecol. 2017;93. DOI: 10.1093/femsec/fix003. [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel Met al. . The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–206. [DOI] [PubMed] [Google Scholar]
- Vašutová M, Mleczko P, López-García Aet al. . Taxi drivers: the role of animals in transporting mycorrhizal fungi. Mycorrhiza. 2019;29:413–34. [DOI] [PubMed] [Google Scholar]
- Venice F, Desirò A, Silva Get al. . The mosaic architecture of NRPS-PKS in the arbuscular mycorrhizal fungus gigaspora margarita shows a domain with bacterial signature. Front Microbiol. 2020a;11:2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venice F, Ghignone S, Fossalunga AS diet al. . At the nexus of three kingdoms: the genome of the mycorrhizal fungus gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ Microbiol. 2020b;22:122–41. [DOI] [PubMed] [Google Scholar]
- Vlk L, Tedersoo L, Antl Tet al. . Alien ectomycorrhizal plants differ in their ability to interact with co-introduced and native ectomycorrhizal fungi in novel sites. ISME J. 2020;14:2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright P, Hinkle G, Sogin Met al. . Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–2. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang Y, Wang Jet al. . Plants acquired a major retrotransposon horizontally from fungi during the conquest of land. New Phytol. 2021. DOI: 10.1111/nph.17568. [DOI] [PubMed] [Google Scholar]
- Warmink JA, Nazir R, Elsas JDV. Universal and species-specific bacterial ‘fungiphiles’ in the mycospheres of different basidiomycetous fungi. Environ Microbiol. 2009;11:300–12. [DOI] [PubMed] [Google Scholar]
- Whiteside MD, Werner GD, Caldas VEet al. . Mycorrhizal fungi respond to resource inequality by moving phosphorus from rich to poor patches across networks. Curr Biol. 2019;29:2043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibbelt G, Kurth A, Hellmann Det al. . White-Nose syndrome fungus (Geomyces destructans) in bats, europe. Emerg Infect Dis. 2010;16:1237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig MR, Kaufman DM, Stevens RD. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu Rev Ecol Evol Syst. 2003;34:273–309. [Google Scholar]
- Worrich A, Stryhanyuk H, Musat Net al. . Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments. Nat Commun. 2017;8:15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Jiang D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol. 2014;52:45–68. [DOI] [PubMed] [Google Scholar]
- Yan JF, Broughton SJ, Yang SLet al. . Do endophytic fungi grow through their hosts systemically?. Fungal Ecol. 2015;13:53–9. [Google Scholar]
- Yildirir G, Malar C M, Kokkoris Vet al. . Parasexual and sexual reproduction in arbuscular mycorrhizal fungi: room for both. Trends Microbiol. 2020;28:517–9. [DOI] [PubMed] [Google Scholar]
- Zanne AE, Abarenkov K, Afkhami MEet al. . Fungal functional ecology: bringing a trait-based approach to plant-associated fungi. Biolog Rev. 2020;95:409–33. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang S, Li Yet al. . Phylogeography and evolution of a fungal–insect association on the tibetan plateau. Mol Ecol. 2014;23:5337–55. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li X, Kotta-Loizou Iet al. . A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 2021;15:1893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]