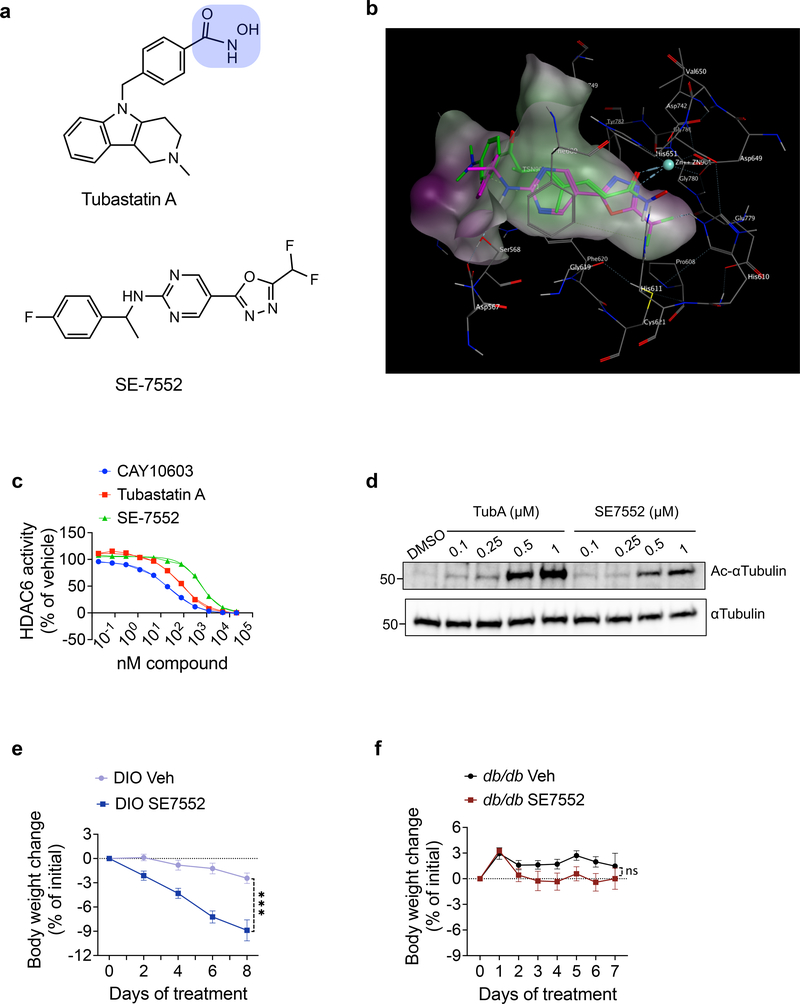
Fig. 8. A non-hydroxamate HDAC6-specific inhibitor reverses diet-induced obesity.
a, Chemical structures of tubastatin (top) and SE-7552 (bottom). The hydroxamate moiety of tubastatin is highlighted in blue. b, Docking of SE-7552 into the inhibitor binding site of HDAC6. The A chain of the 5edu.pdb was used for docking. The Trichostatin A in the 5edu.pdb is shown in green and the docked pose of the SE-7552 in magenta. One nitrogen of oxadiazole in SE-7552 is binding to Zinc as the carbonyl of hydroxamate in Trichostatin A, one of the fluorine in the difluoromethyl group is interacting with His610, the pyrimidine ring is sandwiched between Phe680 at the top and Phe620 at the bottom making pi-pi interactions with them. The amide is hydrogen bonding with Ser568, and the fluorobenzene ring is also interacting with Phe620 by T-shaped pi-pi stacking. The docking experiment was carried out using MOE software 86. The same results were obtained by gold docking program 87. c, Dose response curves of HDAC6 inhibition for the HDAC6-specific inhibitors CAY10603, tubastatin, and SE-7552 (n=2 per dose per compound, n=6 for no compound). d, Immunoblots from lysates of N1 cells treated with the indicated compounds for 24 hr. e, f, Body weight change of SE-7552 (50 mg/kg, i.p., n=12) treated wild-type DIO mice (n=13, P=3E-10 by mixed-effect analysis with Sidak correction) (e) or db/db mice (n=6) (f). *P<0.05, **P<0.01, ***P<0.001. Data are represented as mean ± s.e.m.