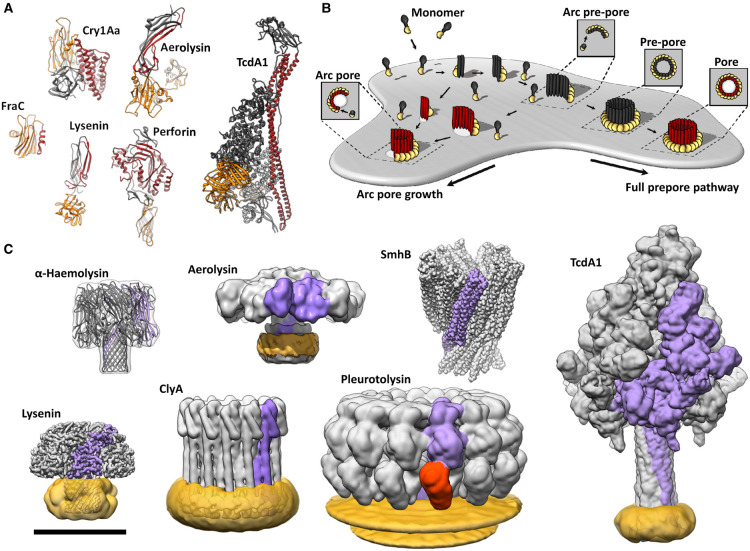
Figure 1. Pore-forming proteins are structurally diverse.
(A) Protomers of lysenin (RCSB Protein Data Bank [PDB] ID code 3ZXG), Bacillus thuringiensis Cry1Aa (PDB: 1CIY), aerolysin (PDB: 3G4N), Fragaceatoxin C (FraC; PDB: 3LIM), perforin (PDB: 3NSJ) and Photorhabdus luminescens Tc holotoxin (TcdA1; PDB: 6RW6). Motifs/domains responsible for membrane insertion and membrane binding are coloured red and orange, respectively. (B) Schematic depicting a generic pathway for pore formation into a lipid membrane based on MACPF/CDC pore-forming systems. Soluble components, typically monomers, bind the membrane. Several monomers subsequently come together to form oligomers. Oligomeric intermediates undergo a conformational change inserting transmembrane amphipathic regions into the lipid bilayer, forming an aqueous channel. Some other PFPs systems oligomerise in solution before binding the bilayer. Membrane penetration may precede oligomerisation in some systems. The stoichiometry of the various intermediates is highly dependent on the pore-forming system. Additional intermediates (not depicted) may also be present. (C) Pore forms (volume rendering in UCSF Chimera) of pleurotolysin (PDB: 4V2T), α-haemolysin (PDB: 3M2L), SmhB (PDB: 7A0G) and ClyA (PDB: 6MRW). Low-pass filtered cryoEM maps of aerolysin (EMDB-8187), lysenin (EMDB-8015) and Photorhabdus luminescens Tc holotoxin (TcdA1; EMDB-10313). A single protomer from each pore is coloured purple (and orange, for the two-component system of pleurotolysin). Where applicable, detergent or lipids are coloured yellow. Scale bar is 100 Å for A and C.