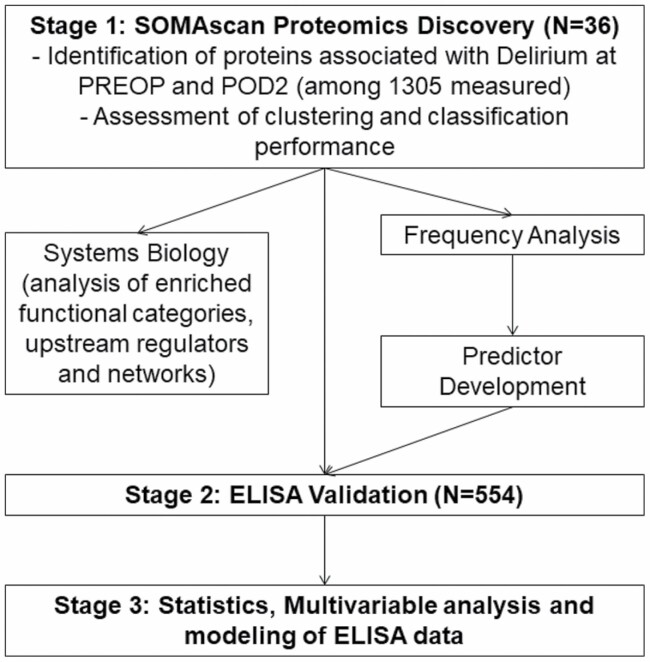
Figure 1.
Overall workflow and methodologic steps for protein identification of delirium protein expression profiles and ELISA validation of proteins. Proteins significantly differentially expressed in delirium samples at PREOP and POD2 were used for hierarchical clustering and PCA to investigate their potential to distinguish the 2 classes of samples. Both PREOP and POD2 delirium-specific protein sets were used for System Biology Analysis to identify their biological implications using Ingenuity Pathway Analysis. Ten thousand subsamples of the data at both PREOP and POD2 were generated to calculate the frequency with which a protein remains delirium-specific. Proteins with high-frequency and fold change, i.e. robust differential expression, were evaluated as predictors of delirium using all possible combinations. For each combination, 1000 4-fold cross-validations were performed to identify predictor performance using support vector machines. ELISA validation of CHI3L1/YKL-40 as the sole delirium-associated protein in both a PREOP and a POD2 predictor model was confirmed by ELISA in a larger cohort of 554 samples. CHI3L1/YKL-40 and markers previously associated with Delirium (CRP [PREOP, POD2] and IL6 [POD2]) were analyzed in a combined model. CHI3L1/YKL-40 = Chitinase-3-like protein 1; CRP = C-reactive protein; ELISA = Enzyme-linked immunosorbent assay; PCA = Principal component analysis; POD2 = Postoperative day 2; PREOP = Preoperative; SVM = Support vector machine.