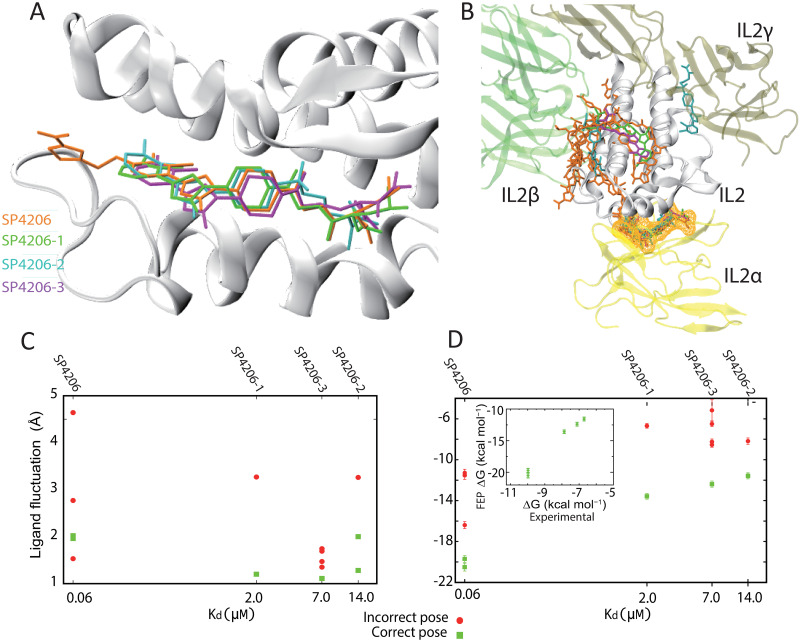
Fig 2. Simulations of binding of SP4206 analog molecules and free energy calculation.
(A) The binding poses of SP4206 and its analogs generated by spontaneous binding simulations. (B) The non-native binding poses of SP4206 and its analogs generated by simulations, shown in the structural context of IL2-IL2R association. The native binding pose is also shown with a mesh around the small molecule. The colors of the small molecules are as shown in (A). (C) The conformational fluctuations of SP4206 and analogs in simulation-generated binding poses (y-axis) compared to the dissociation constants of the compounds (x-axis). As shown, with one exception for SP4206, the fluctuation is smaller in the native binding pose than in the non-native ones for the same compound. (D) The FEP binding free energies of SP4206 and analogs in various simulation-generated binding poses (y-axis) compared to the dissociation constants (Kd) of the compounds (x-axis). The binding free energy was estimated to be 20.5 ± 0.38 or 19.7 ± 0.33 kcal mol−1 for SP4206, 13.58 ± 0.28 kcal mol−1 for SP4206-1, 11.59 ± 0.29 or 11.58 ± 0.28 kcal mol−1 for SP4206-2, and 12.39 ± 0.3 kcal mol−1 for SP4206-3. In the inset, the calculated binding energy (y-axis) is compared to the binding energies derived from Kd (x-axis). For a given compound, the calculated binding energy is consistently greater for the native binding pose than for the non-native ones.