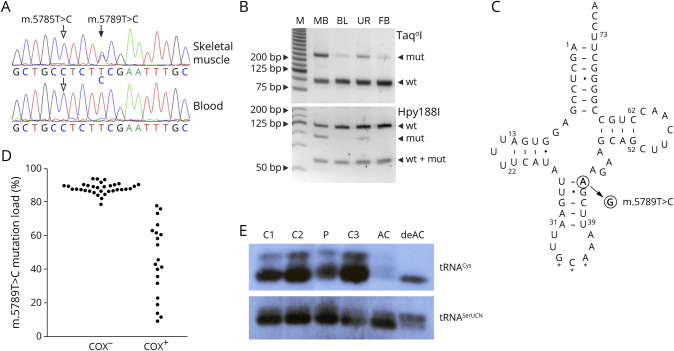
Figure 2. Detection and Functional Effects of the m.5789T>C Sequence Variant.
(A) Sequencing chromatograms from the patient's skeletal muscle and blood. Filled arrow indicates the heteroplasmic m.5789T>C variant, whereas empty arrows show the homoplasmic m.5785T>C variant. (B) Restriction fragment length polymorphism (RFLP) analysis of the patient's skeletal muscle biopsy sample (MB), blood (BL), urine (UR), and fibroblasts (FB). M, molecular weight marker. wt, wild type; mut, mutant. (C) Secondary structure of the human mitochondrial tRNA cysteine adapted from the Mamit-tRNA database.23 Filled arrow indicates the position of the novel m.5789T>C variant. Note that the replacement of a Watson-Crick base pair by a U-G base pair generates 2 adjacent wobble base pairs in the anticodon stem. Stars indicate the anticodon. Dash, Watson-Crick base pair; dot, wobble base pair. (D) Bee swarm diagram of m.5789T>C mutation loads in COX-negative and COX-positive single skeletal muscle fibers as determined by RFLP. Note that fibers with 78% mutation load or less show COX activity. (E) Northern blot analysis of skeletal muscle RNA of the patient (P) and 3 controls (C1–3) as well as aminoacylated (AC) and deaminoacylated (deAC) human osteosarcoma cells 143B RNA. The overall intensity of the tRNACys signal is decreased in comparison to tRNASerUCN, whereas the proportions of aminoacylated and deaminoacylated RNA species are similar in the patient and in controls. COX = cytochrome c oxidase.